Abstract
The Cdc24 protein is essential for the completion of chromosomal DNA replication in fission yeast. Although its precise role in this process is unclear, Cdc24 forms a complex with Dna2, a conserved endonuclease–helicase implicated in the removal of the RNA–DNA primer during Okazaki fragment processing. To gain further insights into Cdc24–Dna2 function, we screened for chromosomal suppressors of the temperature-sensitive cdc24-M38 allele and mapped the suppressing mutations into six complementation groups. Two of these mutations defined genes encoding the Pol3 and Cdc27 subunits of DNA polymerase δ. Sequence analysis revealed that all the suppressing mutations in Cdc27 resulted in truncation of the protein and loss of sequences that included the conserved C-terminal PCNA binding motif, previously shown to play an important role in maximizing enzyme processivity in vitro. Deletion of this motif is shown to be sufficient for suppression of both cdc24-M38 and dna2-C2, a temperature-sensitive allele of dna2+, suggesting that disruption of the interaction between Cdc27 and PCNA renders the activity of the Cdc24–Dna2 complex dispensable.
INTRODUCTION
In all eukaryotic cells, chromosomal DNA replication is achieved by the coordinated action of a large number of protein factors. Replication is a semi-discontinuous process: the leading strand is synthesized continuously, while the lagging strand is synthesized as a series of discrete Okazaki fragments that are later processed to yield a continuous DNA strand. Genetic and biochemical studies in yeast and higher eukaryotic systems, notably the SV40 in vitro replication system, have identified ∼20 polypeptides that are essential for Okazaki fragment synthesis and processing, namely DNA polymerase α-primase (Pol α-primase), replication protein A (RPA), DNA polymerase δ (Pol δ), replication factor C (RFC), the sliding clamp PCNA, the nucleases Fen1 and Dna2, and DNA ligase I (1). All these factors appear to be conserved in all eukaryotic cells.
Current models for Okazaki fragment synthesis and processing can be summarized as follows. First, Pol α-primase synthesizes a short RNA–DNA primer on the template DNA. Then polymerase switching takes place, resulting in displacement of the non-processive low-fidelity Pol α-primase enzyme and its replacement by Pol δ, which is both highly processive when complexed with its processivity factor PCNA and also possesses proofreading activity, ensuring high-fidelity DNA synthesis. Polymerase switching requires the clamp loader complex RFC to recognize the primer-template junction and load the sliding clamp PCNA onto the dsDNA. PCNA encircles the DNA and tethers Pol δ to it. The Pol δ–PCNA complex then continues to synthesize DNA until it encounters the 5′ end of the previously synthesized Okazaki fragment, at which point displacement synthesis results in the formation of an RNA–DNA flap structure. Studies with purified proteins suggest that the extent of displacement synthesis is governed by the binding of the single-stranded binding complex RPA to the displaced flap once the length of the flap has reached ∼35 nt. RPA binding, and its recruitment of the Dna2 helicase–endonuclease, ensures that further displacement synthesis is inhibited. Dna2 then cleaves the flap, most likely removing the entire RNA–DNA segment originally synthesized by the non-proofreading Pol α–primase complex, leaving a shortened flap structure that can be cleaved by the non-essential Fen1 nuclease. The nicked DNA produced is finally sealed by DNA ligase I.
In fission yeast Schizosaccharomyces pombe, Pol δ is composed of four subunits, three of which are essential for cell viability, Pol3, Cdc1 and Cdc27 (2). Orthologues of all four proteins, including the non-essential subunit Cdm1 (3), are found as subunits of mammalian Pol δ (4). Pol3 is the catalytic subunit of the complex. Pol3 binds directly to the B-subunit Cdc1 via a conserved C-terminal zinc finger module (5). The Cdc1 protein then binds to the C-subunit Cdc27 (6). Cdc27 comprises a globular ∼160 residue N-terminal domain (NTD), through which it binds Cdc1, and an extended C-terminal domain of ∼210 residues, at the extreme C-terminus of which is located a PCNA binding motif (PIP box) of the type first identified in the mammalian DNA replication inhibitor p21Cip1 (7). Also located in the C-terminal region of the protein is a conserved, non-essential binding site for Pol1, the catalytic subunit of Pol α–primase (8–10). The D-subunit of the complex, Cdm1, is not essential for cell viability and its role in Pol δ function is not known (3).
In addition to the conserved proteins mentioned above, the fission yeast Cdc24 protein has also been suggested to have a function in lagging strand DNA synthesis. Cdc24 is a 501 amino acid protein with no significant similarity to proteins in current databases (11,12). The involvement of Cdc24 in lagging strand DNA synthesis is suggested from genetic and/or physical interactions with RFC, PCNA, Pol δ and Dna2 (3,11–13). However the precise role of Cdc24 in lagging strand synthesis has not yet been established. In this report, Cdc24 is shown to form a complex with Dna2 in fission yeast protein extracts, extending previous results (13) obtained using the two-hybrid system. We also report the isolation and analysis of cdc24 suppressor mutants. Together with complex formation between Cdc24 and Dna2, we propose that Cdc24 has a role in the processing of flap structures in Okazaki fragment maturation during lagging strand DNA synthesis.
MATERIALS AND METHODS
General fission yeast techniques and reagents
S.pombe genetic techniques and media were as described in reference (14). The S.pombe cDNA library used was described previously (12). The S.pombe genomic DNA libraries were constructed by inserting restriction enzyme digested genomic DNA into plasmid pALSK+.
Epitope tagging
For epitope tagging of cdc24+ and dna2+, plasmid pKSU–XHA was first generated by subcloning the genomic ura4+ HindIII fragment and 3xHA-nmt1 3′ UTR region NotI-SacI fragment from plasmid pREP2–XHA, a version of pREP2 carrying sequences encoding three HA epitopes for C-terminal tagging, into pBluescript II KS+ (Stratagene). Next, cdc24+ genomic DNA was amplified by PCR, digested with EcoRI and NotI (the sites were present in the oligonucleotides used for PCR) and ligated into pKSU–XHA, such that the cdc24+ open reading frame (ORF) was fused in frame at its 3′ end to sequences encoding three copies of the HA mAb epitope. Next, the nmt1 3′ UTR in this plasmid was replaced with the 3′ UTR of cdc24+ by replacing the BamHI-SacI region with a BamHI-SacI cdc24+ fragment generated by PCR, to make pKSU–cdc24HAUTR. For dna2+, plasmid pKSU–dna2HAUTR was constructed in a similar fashion by subcloning dna2+ genomic DNA and dna2+ 3′ UTR sequence into pKSU–XHA. For construction of pKSU–cdc24MycUTR and pKSU–dna2MycUTR, the NotI-BamHI 3xHA fragment in the plasmids described above was replaced with a PCR amplified 13xMyc fragment. All DNA sequences amplified by PCR were confirmed by DNA sequencing. The sequences of the primers used will be provided by the authors on request.
To construct the epitope tagged strains, the pop-in/pop-out homologous recombination method was used (15). The pKSU–cdc24HAUTR and pKSU–cdc24MycUTR plasmids were linearized by HpaI digestion, while pKSU–dna2HAUTR and pKSU–dna2MycUTR plasmids were linearized by BglII digestion. The linear plasmid DNAs were then transformed into ura4-D18 leu1-32 h− cells. After selection of stable ura+ transformants, PCR was used to confirm the correct integration (pop-in) of the plasmids at the desired loci. Ura− cells were then selected on 5-fluoroorotic acid (5-FOA) plates. Loss of the plasmid (pop-out) was confirmed by PCR and followed by immunoblotting analysis. The tagged strains were backcrossed three times and double cdc24 dna2 mutants were obtained by crossing the single mutants.
Immunoprecipitation and immunoblotting
Exponentially growing cells in EMM medium supplemented with uracil and leucine were collected and washed with ice-cold STOP buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA and 1 mM NaN3). Cells were resuspended in Buffer B (10 mM sodium phosphate buffer pH 7.0, 1% Triton X-100, 1mM EDTA and 100mM NaCl) with protease inhibitors, transferred into a tube containing glass beads, and disrupted using a Ribolyser (Hybaid). Cell extracts (2.0 mg or 1.0 mg of proteins in buffer B in panels A and B, respectively) were mixed with anti-HA rat monoclonal antibody (clone 3F10) affinity matrix (Roche) that had been pre-treated with BSA. After incubation for 1 h at 4°C with rotation, the immunoprecipitates were washed four times with buffer B. Crude extracts (20 μg in panel A or 10 μg in panel B) and immunoprecipitates from 2.0 mg or 1.0 mg of extract (as above) were separated by SDS–PAGE, transferred to membrane and detected by anti-HA (clone 12CA5) or anti-Myc (clone 9E10) mouse monoclonal antibodies (Roche).
Classification of cdc24-M38 suppressor mutants
Spontaneous genetic suppressors of cdc24-M38 h− were isolated as described previously (16). Briefly, cdc24-M38 leu1-32 h− cells were incubated at 36°C for 3 days on YE medium. Spontaneous revertant colonies were then replica-plated to 18°C for 3–5 days to test for cold-sensitivity. Following backcrossing, the cold-sensitive suppressor strains were crossed with leu1 h+S to determine whether the suppressing mutations were intra- or extragenic. Subsequently, to group the suppressors into complementation groups, the suppressed cdc24-M38 h− strains were crossed to cdc24-M38 h+S, to allow the isolation of suppressed cdc24-M38 strains of opposite mating type. The different suppressed strains (cdc24-M38 cstX, cdc24-M38 ctsY, etc.) were then crossed with one another and the progeny analysed by random spore analysis. Where all the progeny were cold-sensitive, the suppressing mutations were assigned to the same complementation group. Otherwise, the suppressors were assigned to different complementation groups. In all, the suppressing mutations fell into six complementation groups, designated cst1–cst6.
Construction of cdc27 mutants
Three types of cdc27 deletion mutations were generated by PCR. DNA fragments were digested with EcoRI and NotI, and subcloned into pBluescript II-ura4+ plasmid. The resultant plasmids, pKSU–cdc27-D1, pKSU–cdc27-D2 and pKSU–cdc27-D3, contain 0.5 kb of 3′ UTR derived from the cdc27+ gene and a varying length of coding sequence depending on the length of deletion (see Results). All DNA sequences generated by PCR were confirmed by DNA sequencing. After linearization by PstI digestion, the three plasmids were transformed to ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 h−/h+ diploid cells. Successful homologous recombinants were selected following PCR analysis of genomic DNA. To test whether the cdc27 mutants are viable or not, diploid cells were spread on MEA plates to induce meiosis and sporulation. Spores were then plated on YE medium to allow re-growth, and viable colonies analysed to determine their genotypes. All three cdc27 mutants were viable. These were then plated on 5FOA plates to select for ura− cells, after which PCR analysis was used to confirm the loss of the plasmid sequences. Finally, the cdc27 mutant strains were backcrossed three times with h− leu1-32 or h+ leu1-32 cells to obtain cdc27-D1 leu1-32 h−, cdc27-D2 leu1-32 h− and cdc27-D3 leu1-32 h− mutants for further phenotypic analysis.
Forward mutation rate assay
Wild-type control (leu1-32 h+S) and cdc24-M38 strains (cdc24-M38 leu1-32 h+S) were streaked to single colonies on EMMGL plates at 28°C and eight single colonies of each strain were inoculated into 10 ml of EMMGL medium (EMM with 5 g/l of glutamic acid replacing ammonium chloride, supplemented with 150 mg/l of leucine) and grown to saturation at 28°C for 48 h. From each saturated culture, 1 × 107 cells per plate were plated onto EMMGL agar containing either 40 μg/ml of filter-sterilized L-canavanine sulphate (Sigma) or thialysine (S-2-aminomethyl-L-cysteine hydrochloride, Sigma). The plates were then incubated at 28°C for 8 days and the number of L-canavanine- or thialysine-resistant clones counted. Plating efficiencies were determined by plating 1000 cells of each strain onto EMMGL plates.
RESULTS
Cdc24 and Dna2 form a complex
The fission yeast Dna2 and Cdc24 proteins are essential for the completion of DNA replication (11,12). In the course of previous analysis of fission yeast Dna2, an interaction between Cdc24 and Dna2 was observed using a two-hybrid assay, in which the two proteins were expressed to a high level in Saccharomyces cerevisiae as Gal4 activation domain and Gal4 DNA binding domain fusions (13). To determine whether Cdc24 and Dna2 also formed a complex in vivo in S.pombe when present at their normal cellular levels, fission yeast strains were constructed, in which the endogenous cdc24+ and dna2+ genes were precisely replaced with the alleles encoding epitope tagged versions of the two proteins (see Materials and Methods), thereby ensuring that the tagged proteins were expressed to a normal level only.
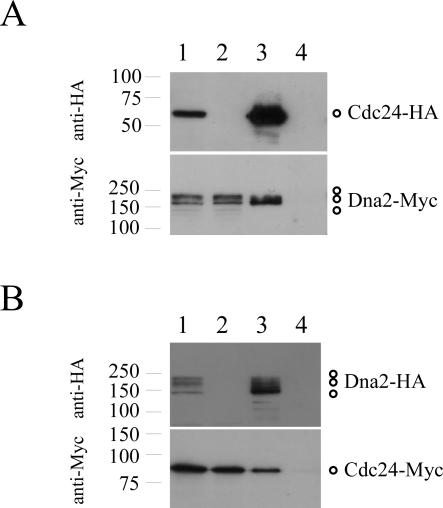
To test whether Cdc24 and Dna2 were physically associated, co-immunoprecipitation assays were performed. Initially, protein extracts were prepared from dna2-Myc cdc24-HA and dna2-Myc strains, and subjected to immunoprecipitation using anti-HA antibodies. The immunoprecipitated material was then resolved on SDS–PAGE and immunoblotted using anti-HA and anti-Myc antibodies. Figure 1A shows the results of these experiments: Dna2-Myc could be immunoprecipitated by the anti-HA antibody but only in the presence of Cdc24-HA (compare lanes 3 and 4), that is, only from extracts prepared from the dna2-Myc cdc24-HA strain (Figure 1A).
Figure 1.
Cdc24 and Dna2 form a complex in vivo. (A) Dna2-Myc co-purifies with Cdc24-HA. Native protein extracts were prepared from cdc24-HA dna2-Myc ura4-D18 leu1-32 h− (lanes 1 and 3) and dna2-Myc ura4-D18 leu1-32 h+ (lanes 2 and 4) cells growing exponentially in EMM medium supplemented with leucine and uracil, and Cdc24-HA immunoprecipitated with anti-HA antibody. Cell extracts (lanes 1 and 2) and immunoprecipitates (lanes 3 and 4) were analysed by immunoblotting with anti-HA (upper panel) and anti-Myc (lower panel) antibodies. (B) Cdc24-Myc co-purifies with Dna2-HA. The same procedures were performed with dna2-HA cdc24-Myc ura4-D18 leu1-32 h− (lanes 1 and 3) and cdc24-Myc ura4-D18 leu1-32 h+ (lanes 2 and 4) cells, but in this case, Dna2-HA was immunoprecipitated with anti-HA antibody. Cell extracts (lanes 1 and 2) and immunoprecipitates (lanes 3 and 4) were then analysed by immunoblotting with anti-HA (upper panel) and anti-Myc (lower panel) antibodies. Molecular weight markers (kD) are shown to the left of each panel. Note that multiple forms of Dna2 can be detected by immunoblotting. Whether these represent post-translationally modified isoforms of the protein, or merely degradation products, is not known.
This result was confirmed with another combination of tags. Native proteins extracts were prepared from dna2-HA cdc24-Myc and cdc24-Myc strains. After immunoprecipitation using anti-HA antibody and immunoblotting, we found that Cdc24-Myc was specifically immunoprecipitated by anti-HA antibody only in the presence of Dna2-HA, that is, only from extracts prepared from the dna2-HA cdc24-Myc strain (Figure 1B, compare lanes 3 and 4). These results indicate that Cdc24 forms a complex with Dna2 in yeast protein extracts, and therefore, most likely in vivo also, and suggests that, like Dna2, Cdc24 functions in Okazaki fragment maturation.
Complementation groups of cdc24-M38 suppressing mutations
In order to investigate the function of cdc24+ further, and in particular, to question its potential roles during Okazaki fragment synthesis and maturation, extragenic suppressors of the temperature-sensitive cdc24-M38 allele were isolated as has been previously described (16). Briefly, exponentially growing temperature-sensitive cdc24-M38 cells were spread onto YE plates and incubated at the restrictive temperature of 36°C for 3 days, in order to allow the growth of spontaneous revertant colonies. In order to facilitate subsequent analyses, cold-sensitive revertants were then selected following replica plating and incubation at 18°C for 3–5 days. The suppressing mutants were designated cst mutants, for cold-sensitive suppressors of twenty-four. Note that the cst mutations confer two properties on cells: cold-sensitivity and suppression of the temperature-sensitivity of cdc24-M38.
In total, 37 such mutants were isolated from ∼3 × 109 plated cells. Subsequent genetic analyses showed that all the cst mutant phenotypes were caused by single gene mutations and not by reversion of the cdc24-M38 allele to wild type. To classify the cst mutants into complementation groups, the mutants were crossed to one another and the resulting spores were plated and incubated at 18 and 36°C. Where all the progeny were cold-sensitive, the suppressing mutations were assigned to the same complementation group. Otherwise, the suppressors were assigned to different groups. In all, the 37 mutants were classified into six complementation groups, cst1–cst6, containing between 1 and 17 alleles (Table 1). One of these complementation groups, cst6+ has been described (16,17). The cst6+ gene, which encodes an essential DNA helicase enzyme, was renamed pfh1+.
Table 1. Complementation groups of cdc24 suppressing mutants.
| No. of alleles | Morphologya | Gene | Encoded protein | |
|---|---|---|---|---|
| cst1+ | 17 | Elongated | cdc27+ | Pol δ subunit |
| cst2+ | 4 | Normal | sup45+ | eRF1 |
| cst3+ | 8 | Normal | sup35+ | eRF3 |
| cst4+ | 2 | Elongated | pol3+ | Pol δ subunit |
| cst5+ | 1 | Elongated | – | – |
| cst6+ | 5 | Elongated | pfh1+ | DNA helicase |
aCell morphology of double mutant with cdc24-M38 at 18°C.
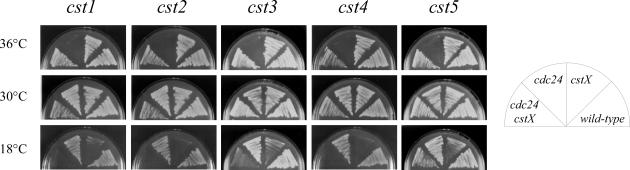
Figure 2 shows representative alleles from the cst1–cst5 complementation groups. The mutant cells show cold-sensitivity, in the presence and absence of cdc24-M38, as well as the suppression of the temperature-sensitivity of cdc24-M38. Microscopic observation of cst mutants incubated at 18°C revealed that they could be divided into two groups. Mutants in the first group displayed an elongated morphology at 18°C. This group consisted of mutants of cst1+, cst4+, cst5+ and cst6+/pfh1+. In contrast, the second group of mutants, comprising alleles of cst2+ and cst3+, did not show the elongated cell phenotype. The cell elongation phenotype suggested that the cst1+, cst4+, cst5+ and cst6+/pfh1 genes could be involved in cell cycle events (Table 1).
Figure 2.
Suppression of the temperature-sensitivity of cdc24-M38 by the cst1-R3, cst2-R11, cst3-R13, cst4-R18 and cst5-R19 mutations. Cells from the indicated strains were streaked on YE plates and incubated for 3 days at 36°C or 30°C, or 5 days at 18°C.
Identification of cst genes
To identify the cst genes, two approaches were taken. The first was to examine whether the cold-sensitive phenotype of the cst mutants could be rescued by the introduction of known genes. Previously, a variety of genetic interactions between cdc24 and genes encoding components of the Pol δ-RFC-PCNA holoenzyme have been reported. For example, overproduction of PCNA will partially rescue cdc24-M38 (3,12), as will overproduction of Rfc1, the large subunit of the RFC complex (12). Six candidate genes were therefore tested, for their ability to complement the cold-sensitivity of individual cst or cdc24 cst double mutants, when expressed from multi-copy plasmids. The genes examined were pol3+/cdc6+, cdc1+ and cdc27+ (encoding the catalytic, B and C subunits of Pol δ, respectively), cdc17+ (DNA ligase I), pcn1+ (PCNA) and rfc1+ (encoding the large subunit of RF-C). Among these genes, two were observed to efficiently rescue particular cst mutants: pol3+ rescued cst4 and cdc27+ rescued cst1 (data not shown). To test whether pol3+ was isogenic with cst4+, genetic linkage between these loci was analysed by crossing pol3/cdc6 and cst4 mutants. The results of these experiments showed that pol3+ and cst4+ are tightly linked, implying that cst4+ is pol3+. By similar analysis, it was also concluded that cst1+ is isogenic with cdc27+. Thus, mutations in genes encoding two different subunits of Pol δ were identified as cst mutations. This strongly suggests a physiological interaction between Cdc24-Dna2 and Pol δ.
In contrast, none of the six candidate genes suppressed the cold-sensitivity of cst2, cst3 and cst5 mutants. Therefore, genomic and cDNA libraries were screened for genes that suppressed the cold-sensitivity of these mutants. No suppressors of cst5 were isolated, and the identity of this gene remains unknown. A single gene, sup45+, was identified as a suppressor of cst2-R11. This gene (systematic name SPAC1834.01, see http://www.sanger.ac.uk/Projects/S_pombe for details) was isolated from both cDNA and HindIII digested genomic DNA libraries and encodes the translation termination factor eRF3. In vivo, eRF3 forms a complex with eRF1, which is encoded by sup35+ (18). Strikingly, sup35+ (SPCC584.04) was identified as the only suppressor of cst3-R13. A single HindIII DNA fragment was isolated from genomic DNA libraries as a cst3-R13 suppressor and subsequent deletion analysis revealed that the complementation activity in this plasmid was derived from the sup35+ gene (data not shown). These results suggest that cst2+ and cst3+ correspond to the fission yeast sup45+ and sup35+ genes, respectively. To examine this, the sup45+ and sup35+ genes in the genomes of cst2-R11, cst3-R13 and wild-type cells were sequenced. A single point mutation in sup45 was found in cst2-R11 but not in cst3-R13 or wild-type strains. The cst2-R11 allele contains a single C to A (CAA to AAA) point mutation, resulting in a single amino acid change, from proline to lysine, at residue 400 in the predicted Cst2/Sup45-R11 protein. Similarly, a point mutation in sup35 gene was only found in cst3-R13 strain. The cst3-R13 allele contains a single C to A (GAC to GAA) point mutation, resulting in a single amino acid change, from aspartic acid to glutamic acid, at residue 292 in the predicted Cst3/Sup35-R13 protein. Therefore, cst2+ and cst3+ are indeed sup45+ and sup35+, respectively. Since the molecular defect in the cdc24-M38 allele is a nonsense mutation at codon 370, it is probable that this mutant can be suppressed by mutations that suppress premature translational termination at this codon. This interpretation is consistent with our observation that neither cst2 nor cst3 mutants displayed an elongated cell phenotype (Table 1), suggesting that the mutants do not have a cell cycle phenotype.
Sequencing of pol3 and cdc27 mutant alleles
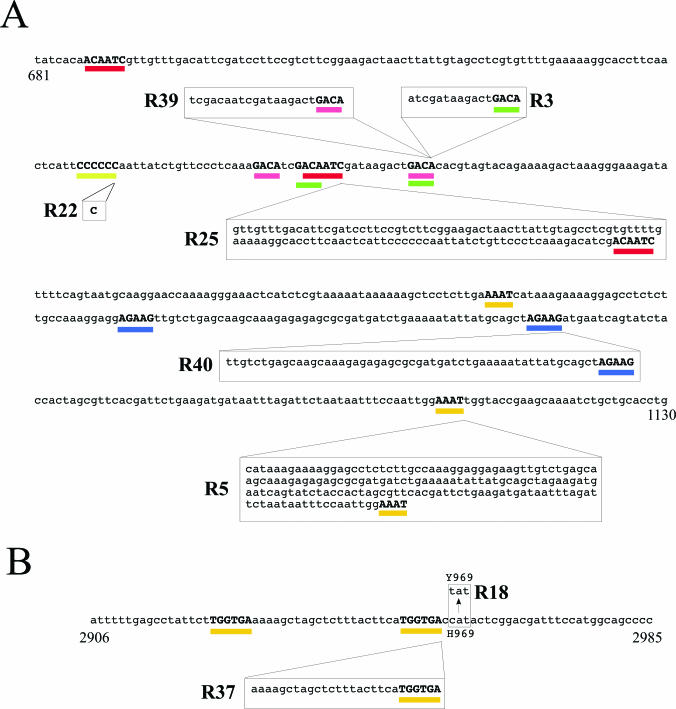
To gain insights into the type of mutations obtained and the effect of these mutations on the mutant proteins, all the cdc27 and pol3 mutant alleles were amplified by PCR and the PCR products were directly sequenced. Among the seventeen cdc27 alleles, we found six different mutations (Figure 3, Table 2). Eight of the mutant alleles (cdc27-R22, etc.) contained a single nucleotide insertion of a cytosine residue in a stretch of six consecutive cytosines (Figure 3A, indicated by yellow bar). This frame-shift mutation causes truncation of the native Cdc27 protein after amino acid 164, although the mutant protein gains some extra 12 amino acids derived from another reading frame before terminating. All the other cdc27 mutant alleles contained sequence duplications ranging in size from 16 to 173 bp. Each duplicated sequence is flanked by a short direct repeat, 4–7 nt in length, with no apparent sequence specificity. The cdc27-R3 allele has 16 bp duplication between two GACA sequences (Figure 3A, green bars), while cdc27-R39 allele has 22 bp duplication between GACA sequences (Figure 3A, pink bars). One of the GACA sequence surrounding the duplicated sequence in cdc27-R39 is identical to that of cdc27-R3 (Figure 3A). Of the remaining alleles, cdc27-R40 has 58 bp duplication between AGAAG sequences (Figure 3A, blue bars), cdc27-R25 has 121 bp duplication between ACAATCG sequences (red bars) and cdc27-R5 has a 173 bp duplication between AAAT repeats (orange bars). All these sequence duplications result in truncation of the C-terminal region of the Cdc27 protein due to frame-shift. Table 2 summarizes these results and the predicted lengths of the mutant proteins.
Figure 3.
Mutation spectra for cst alleles of cdc27 and pol3. (A) Nucleotide sequence of a 450 bp region of cdc27+ genomic DNA (from nucleotide 681–1130, where the ATG is at 1) showing the molecular structure of the cdc27 cst alleles. Six different mutation types are indicated, as follows: found in R39 allele (22 bp duplication of region bounded by GACA repeats underlined in pink), R3 and three isoalleles (16 bp duplication, GACA repeats, green), R25 and one isoallele (121bp, ACAATCG, red), R40 allele (58 bp, AGAAG, blue), R5 allele (173 bp, AAAT, orange) and R6 and seven isoalleles (insertion of single C in CCCCCC sequence, indicated by yellow bar). (B) Nucleotide sequence of an 80 bp region of pol3+ genomic DNA (from nucleotide 2906–2985, where the ATG is at 1) showing the molecular structure of the pol3 cst alleles. Two different mutants were obtained: R37 (27 bp duplication of region bounded by TGGTGA repeats underlined in orange), and R18 (C to T mutation, causing H969 to be mutated to Y969).
Table 2. Summary of cdc27 mutant alleles.
| Mutation type | Repeated sequence | Duplication size | Occurrence | Mutant protein lengtha |
|---|---|---|---|---|
| Insertionb | – | – | 8/17 | 164 + 12 |
| Duplication | GACA | 16 | 4/17 | 179 + 2 |
| Duplication | GACA | 22 | 1/17 | 179 + 4 |
| Duplication | AGAAG | 58 | 1/17 | 245 + 5 |
| Duplication | ACAATCG | 121 | 2/17 | 175 + 2 |
| Duplication | AAAT | 173 | 1/17 | 271 + 24 |
aThe length of the mutant protein is shown as number of amino acids from Cdc27 + number of additional amino acids resulting from frame-shift.
bThe insertion alleles contained one additional C residue inserted into a run of six C residues.
The two cold-sensitive pol3 alleles were also sequenced. The pol3-R18 allele contains a single C to T point mutation, resulting in a single amino acid change, from histidine to tyrosine, at residue 969 in the Pol3-R18 protein. In pol3-R37 allele, there is a 27 bp sequence duplication (Figure 3B, orange bars). This duplication also occurred between short repeat sequences (TGGTGA) and is located only a few bases from the position of the pol3-R18 mutation (Figure 4). The duplication causes an in-frame nine amino acid insertion (sequence EKASSLLHG) into the Pol3 protein sequence between amino acid 967 and 968. Both mutations are located in a region of the Pol3 protein of unknown function that lies between the catalytic (polymerase) domain and the tandem C-terminal zinc finger modules responsible for the interaction between Pol3 and the B-subunit of the Pol δ complex, Cdc1 (5).
Figure 4.
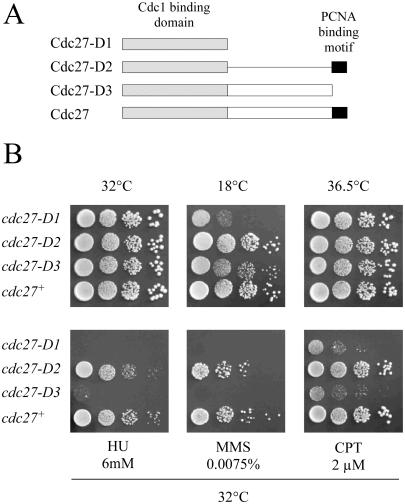
Properties of truncated Cdc27 proteins. (A) Schematic representation of wild-type and C-terminally truncated Cdc27 mutant proteins, showing the location and extent of the globular Cdc1 binding domain (speckled box) and the PCNA binding motif (black box). Cdc27-D1 lacks amino acids 160–372; Cdc27-D2 lacks 160–352; Cdc27 lacks 353–372. (B) Growth properties and drug sensitivities of cdc27-D1, cdc27-D2 and cdc27-D3 mutant strains at 32°C. Serial dilutions of exponentially growing cdc27-D1, cdc27-D2, cdc27-D3 and wild-type cells in YE were spotted (10 000, 1000, 100, 10 cells per spot) onto YE plates or YE plates containing 6mM HU, 0.0075% MMS and 2 μM CPT. Plates at 32°C and 36.5°C were incubated for 3 days. The 18°C plate was incubated for 7 days.
Phenotype of Cdc27 C-terminal deletion mutants
The Cdc27 protein is essential for complete DNA replication in fission yeast (19). Biochemical studies have shown that the 372 amino acid protein has a globular N-terminal domain of ∼160 amino acids that binds to Cdc1, and a highly extended C-terminal region (6,20,21). There is a binding site for PCNA at the extreme C-terminus of the protein (amino acids 363–372) (20). Also within the C-terminal region is a short conserved sequence motif that binds to the catalytic subunit of the Pol α-primase complex (10). Previous studies have suggested that the C-terminal region of the Cdc27 protein is essential for its in vivo function (20,21). Deletion of the last 20 amino acids of Cdc27 has been shown to render the protein unable to complement a cdc27 allele when the truncated mutant protein is expressed from a heterologous promoter on a multi-copy plasmid (20,21). In vitro, loss of the PCNA binding site greatly reduces the processivity of the S.pombe Pol δ enzyme (21).
Despite the apparent importance of PCNA binding for Cdc27 function, the identification of truncated Cdc27 proteins that act as suppressors of cdc24-M38 indicates that, when the mutant proteins are expressed from the endogenous cdc27+ promoter, the PCNA binding motif is non-essential. To confirm this (and to rule out the possibility that sequences encoding the PCNA binding motif might be restored to the 3′ end of the mutated cdc27 ORF by some aberrant splicing event), three new mutant alleles were constructed by replacing the endogenous cdc27+ gene with truncated gene sequences (Figure 4A). Of the encoded proteins, Cdc27-D1 possesses only the N-terminal Cdc1 binding domain (160–372), Cdc27-D2 lacks most of the of C-terminal region but retains the PCNA binding motif (160–352), while Cdc27-D3 lacks only the PCNA binding motif (353–372). By using a two-step homologous recombination method in diploid cells, heterozygous mutant strains were obtained that contained, on one chromosome, the mutant genes under the control of the native cdc27 promoter. The diploids were then induced to sporulate, the haploid spores were allowed to germinate on rich medium, and the genotypes of the resulting clones analysed by PCR.
All three haploid cdc27-D mutants were viable at 32°C. The mutant strains were then examined for temperature, cold or drug sensitivities (Figure 4B). All three mutants grew well at 32°C. However, both cdc27-D1 and cdc27-D3 showed cold-sensitivity; cdc27-D3 cells could grow only slowly at 18°C, and cdc27-D1, not at all. Cells of both mutants were highly elongated at 18°C, indicating cell cycle delay. In addition to this, cdc27-D1 and cdc27-D3 cells were highly sensitive to the ribonucleotide reductase inhibitor hydroxyurea (HU), the DNA damaging agent methyl methanesulphonate (MMS) and the topoisomerase I inhibitor camptothecin (CPT). These results indicated that, while the PCNA binding motif at the C-terminus of Cdc27 is not absolutely required for cell growth, it is required for the efficient growth at low temperature and for cell viability after DNA damage.
Loss of PCNA binding domain suppresses cdc24-M38
Six different cdc27 alleles were identified as suppressors of cdc24-M38, all of which appear to encode truncated proteins (Table 2). The longest protein encoded by the mutant alleles, Cdc27-R5, comprises 271 amino acids of Cdc27 sequence, plus an additional 24 amino acids that result from the frame-shift caused by the sequence duplication. Therefore, loss of 101 amino acids from the extended C-terminal part of Cdc27 appears to be sufficient for cdc24-M38 suppression activity. This region of the protein contains binding sites for Pol1 (amino acids 293–332) and for PCNA (amino acids 353–372) (10,20).
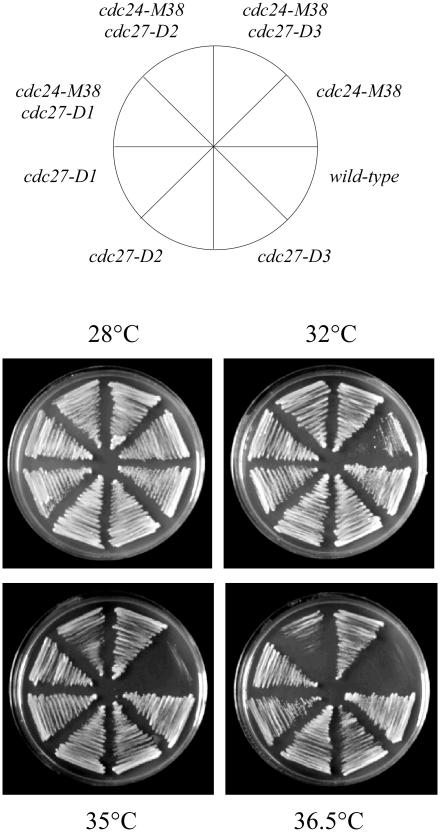
To test whether loss of the PCNA binding motif alone was sufficient for suppression of cdc24-M38, the double mutants cdc24-M38 cdc27-D1, cdc24-M38 cdc27-D2 and cdc24-M38 cdc27-D3 were constructed by tetrad dissection and examined for temperature-sensitivity at 36.5°C (Figure 5). Both cdc24-M38 cdc27-D1 and cdc24-M38 cdc27-D3 cells were able to grow efficiently even at 36.5°C. The Cdc27-D1 protein lacks the entire C-terminal region of the protein, while Cdc27-D3 protein lacks the C-terminal 20 amino acids, containing PCNA binding domain only. Thus, loss of the PCNA binding domain alone is sufficient to restore growth to cdc24-M38 cells at the restrictive temperature. Interestingly, although cdc24-M38 cdc27-D2 cells were able to grow at 35°C, their growth was poor compared with either cdc24-M38 cdc27-D1 or cdc24-M38 cdc27-D3 cells (Figure 5), indicating that the presence of the PCNA binding motif prevents efficient rescue of cdc24-M38. This is discussed further below.
Figure 5.
The cdc27-D1 and cdc27-D3 mutations suppress the temperature-sensitivity of cdc24-M38 cells. cdc24-M38 cdc27-D1, cdc24-M38 cdc27-D2 and cdc24-M38 cdc27-D3 double mutants, plus single mutant and wild-type controls were streaked on YE plates and incubated at the indicated temperatures for 2 days.
The pol3 and cdc27 mutations also suppress dna2-C2
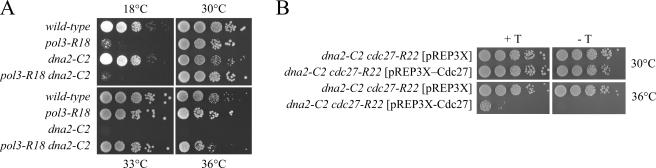
The temperature-sensitive cdc24-M38 and dna2-C2 mutants show similar phenotypes. Cells carrying either mutant allele arrest the cell cycle in S phase with incompletely replicated chromosomes when shifted to the restrictive temperature (12,13,19). As shown above (Figure 1), the Dna2 and Cdc24 proteins form a complex in the cell. These data raised the possibility that the molecular defects of both mutants are the same. If this was the case, the dna2-C2 mutant might be expected to be suppressed by the pol3 and cdc27 alleles that suppress the cdc24-M38. To assess this possibility, dna2-C2 pol3-R18 and dna2-C2 cdc27-R22 mutants were constructed and examined for temperature-sensitivity at 36.5°C (Figure 6). Both pol3-R18 and cdc27-R22 suppressed the temperature-sensitivity of dna2-C2 mutant. This genetic result suggests that Cdc24 and Dna2 have a common function that can be bypassed by impairing the PCNA binding capability of Pol δ.
Figure 6.
The pol3 and cdc27 mutations suppress dna2-C2. (A) The pol3-R18 mutation suppresses the temperature-sensitivity of dna2-C2. Cells from the indicated strains were spotted in 10-fold dilutions (105, 104, 103, 102 and 101 cells) onto YE plates and incubated for 3 days at 36°C, 33°C or 30°C, or at 18°C for 6 days. (B) The cdc27-R22 mutation suppresses the temperature-sensitivity of dna2-C2 and the expression of wild-type cdc27+ abolishes the suppression. The dna2-C2 cdc27-R22 double mutant cells was transformed with pREP3X alone or pREP3X-cdc27 expressing wild-type cdc27+. Each transformant was grown in EMM containing thiamine (5 μg/ml) and the cells obtained were spotted in 10-fold dilutions (105, 104, 103, 102 and 101 cells) onto EMM plates or EMM plate containing thiamine (5 μg/ml). The plates were incubated for 3 days at 36°C or 30°C.
Forward mutation assay
Yeast strains carrying mutations in genes encoding important replicative enzymes often display elevated mutation rates when grown under permissive or semi-permissive conditions and the nature of the mutations that arise can be a useful indicator of the function of the protein in wild-type cells. In fission yeast, for example, inactivation of the Fen1 (S.pombe Rad2) nuclease results in a 16.5-fold increase in mutation rate over wild type, with many of the mutations being sequence duplications flanked by short direct repeat sequences (22), similar in nature to those seen in the pol3 and cdc27 alleles isolated as suppressors of cdc24-M38 (Figure 3, Table 2). To test whether partial impairment of Cdc24 function also resulted in an elevated mutation rate, wild-type and cdc24-M38 cells were plated at 1 × 107 cells per plate onto plates containing 40 μg/ml of either the toxic arginine analogues L-canavanine sulphate (23) or thialysine (S-2-aminoethyl-L-cysteine hydrochloride) (24) and incubated at 28°C for 8 days (see Materials and Methods). Resistance to canavanine or thialysine in fission yeast results from mutational inactivation of can1+, a gene that most likely encodes a regulator of arginine uptake (S. A. MacNeill, unpublished observations) rather than an arginine permease as originally proposed (23). No significant differences were observed between wild-type and cdc24-M38 strains in this system, however, suggesting that cdc24-M38 does not have a mutator phenotype at 28°C. Note that the choice of 28°C for these assays was dictated by the fact that, at higher temperatures, the plating efficiency of cdc24-M38 (in the absence of either arginine analogue) dropped markedly in comparison to wild type.
DISCUSSION
Cdc24 is essential for DNA replication in fission yeast. Cells carrying temperature-sensitive mutations in the cdc24+ gene undergo cell cycle arrest when shifted to the restrictive temperature, becoming arrested with incompletely replicated chromosomes (11,12,19). In this paper, it is shown that Cdc24 interacts with Dna2 in yeast protein extracts, confirming previous results obtained using the two-hybrid system (13). Taken together, these results strongly support a model in which Dna2 and Cdc24 form a functional complex in vivo. Dna2 is a helicase–nuclease that has been implicated in Okazaki fragment processing through a series of genetic and biochemical studies in budding and fission yeast (13,25–32). Dna2 is an essential protein that is conserved throughout eukaryotes. In contrast, homologues of Cdc24 have not been identified in other species, raising the possibility that the protein is unique to S.pombe. In budding yeast, the NTD of the Dna2 protein has been shown to have a regulatory function that is mediated via its interaction with the nuclease and helicase domains for the protein (33). Whether the N-terminus of the fission yeast Dna2 protein has a similar function or not is not clear. However, it is this region of Dna2 that is responsible for binding to Cdc24, as a C-terminally truncated form of Dna2 comprising the N-terminal 425 amino acids only, Dna2-C425, is able to bind Cdc24 in the two-hybrid system (J. Crossland and S. M., unpublished). Therefore, Cdc24 may have a role in regulating Dna2 activity via the Dna2 N-terminal domain. However, confirmation of this will require detailed biochemical analysis of Cdc24–Dna2 interactions. Intriguingly, Dna2 also appears to exist in several forms of differing mobility on SDS–PAGE (see Figure 1). Budding yeast Dna2 and probably the fission yeast Dna2 is phosphorylated by both the Rad53 and Cdc7-Dbf4 kinases (Y. -S. S, unpublished data). In the co-precipitation experiments shown in Figure 1, Cdc24 appears to bind preferentially to the faster migrating forms of Dna2, perhaps hinting at a role for phosphorylation in regulating the Dna2–Cdc24 interaction. Further analysis of this issue is underway.
Through suppressor mutant screening, genetic interactions have been uncovered between cdc24 and genes encoding two of the four subunits of Pol δ, the catalytic subunit Pol3 and the C-subunit Cdc27 (Table 1). Previously, it was shown that overproduction of the D-subunit of Pol δ, Cdm1, was also able to suppress the cdc24-M38 and cdc24-C26 alleles (3). These results suggest that Cdc24 is intimately involved with Pol δ in lagging strand DNA synthesis, consistent with the observation that Cdc24 interacts biochemically with both RFC and PCNA (12).
Sequencing of the cdc27 and pol3 alleles that suppressed cdc24-M38 revealed that many of the mutations (10 out of a total of 19) were DNA sequence duplications (Figure 3, Table 2). All the cdc27 alleles, both sequence duplications and insertions, encoded C-terminally truncated proteins lacking the PCNA binding motif. Indeed, loss of the PCNA binding domain was sufficient to suppress the temperature-sensitivity of cdc24-M38 (Figure 5). Although loss of the PCNA binding motif could result from either sequence insertions or deletion, it was striking that no deletions were found. Furthermore, all the sequence duplications were flanked by short direct repeats. This pattern of mutation has been observed previously in Fen1 mutants in both budding (34) and fission yeast (22). The nuclease Fen1 also plays an important, albeit non-essential, role in Okazaki fragment processing (35) and Fen1 mutant cells (rad27 in S.cerevisiae, rad2 in S.pombe) are thought to accumulate uncleaved flap structures generated during Okazaki fragment processing. Repeat slippages occurring in such a situation are thought to cause duplication mutations. Thus, the propensity for this type of mutation to arise in cdc24-M38 cells suggests that these cells too have a defect in Okazaki fragment processing. Given the close functional and physical relationship between Cdc24 and Dna2, and the phenotypic similarity between mutants in the two genes, it is to be expected that sequence duplications will also be found with increased frequency in S.pombe dna2 mutants also, but this has not been tested. In addition, it is unclear whether sequence duplications will be the predominant type of mutation found in cdc24-M38 cells or whether the selective pressure imposed in the search for suppressors of cdc24-M38 that are able to grow at 36°C resulted in a bias towards this type of mutation.
In the course of this study, it was shown that the PCNA binding domain of Cdc27 is not essential for cell viability, contrary to previous reports (20,36). The reason for this discrepancy is unclear but could relate to the level of expression of the mutant proteins, as in both previous sets of experiments, the mutant proteins were expressed from cloned cDNAs under the control of various forms of the heterologous nmt promoter on multi-copy plasmids (20,36). To analyse the function of the C-terminal part of Cdc27 that includes the PCNA binding domain, we generated three new cdc27 mutants, cdc27-D1, cdc27-D2 and cdc27-D3 (Figure 4A). Both cdc27-D1 and cdc27-D3 cells showed cold-sensitivity and sensitivities to the DNA damaging agents HU, MMS and CPT (Figure 4B). The Cdc27-D1 and Cdc27-D3 proteins both lack the PCNA binding motif. PCNA binding by Cdc27 is therefore essential for growth at low temperature and viability after exposure to certain DNA damaging agents. In contrast to cdc27-D1 and cdc27-D3, cdc27-D2 encodes a protein that lacks most of the C-terminal region (amino acids 160–352) but retains the PCNA binding motif. Cells expressing the Cdc27-D2 mutant protein, unlike those expressing either Cdc27-D1 or Cdc27-D3, were neither cold-sensitive nor sensitive to DNA damaging agents (Figure 4B). Similarly Cdc27-D2 failed to suppress cdc24-M38 efficiently at 36.5°C, although some suppression was apparent at 35°C, suggesting that simply fusing the PCNA binding motif to the globular N-terminal domain of Cdc27 may not completely restore Cdc27 function to wild-type levels. Whether this is due to the Cdc27–PCNA interaction being less than optimal in cdc27-D2 cells or whether the loss of amino acids 160–352 disrupts another important protein–protein interaction (10) awaits biochemical investigation. In addition, note that although interaction with PCNA is essential for the processivity of the Pol δ enzyme, Pol δ interacts with PCNA in at least two ways—via Cdc27 and via the Pol3 and/or Cdc1. Thus, disruption of the Cdc27–PCNA interaction does not abolish Pol δ–PCNA interactions altogether, but is likely to have more subtle effects on enzyme function that are manifest in the cold-sensitivity, drug sensitivity and cdc24-M38 suppression phenotypes displayed by cdc27-D1 and cdc27-D3.
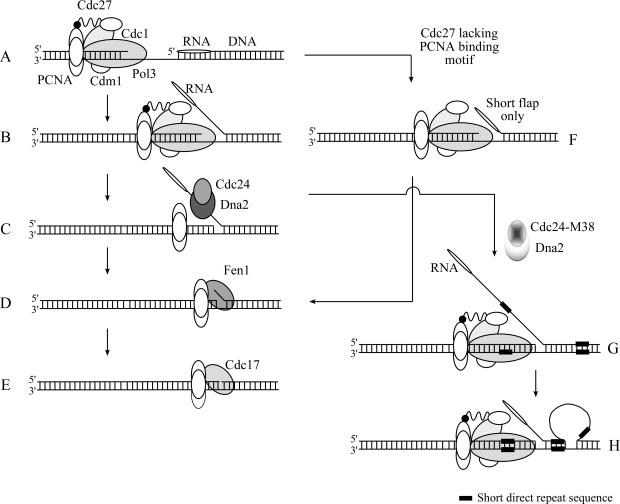
The left side of Figure 7 shows a model for Okazaki fragment processing in fission yeast. Under normal circumstances, maximal processivity of Pol δ is ensured through interactions between Cdc27 and PCNA, and between Pol3 and/or Cdc1 and PCNA. As the polymerase approaches the 5′ end of the previously synthesized Okazaki fragment (Figure 7A), synthesis continues until a flap structure of at least 35 nt is displaced (Figure 7B and C). At this point, the single-stranded DNA binding factor RPA binds the displaced flap and recruits the Dna2–Cdc24 complex. (For clarity, RPA is omitted from the diagram.) Cdc24 may serve to regulate Dna2 or perhaps to link Dna2 to other components of the replication apparatus. Dna2 then cleaves the bulk of the flap, leaving behind a shorter flap that acts as a substrate for the non-essential nuclease Fen1 (Figure 7D). Finally, the single-stranded nick is ligated by DNA ligase I, Cdc17 (Figure 7E). Both Cdc24 and Dna2 are essential proteins, implying that the efficient removal of long flap structures is necessary for successful replication. When the processivity of Pol δ is reduced by disruption of the Cdc27–PCNA interaction (notice the loss of the C-terminal region of Cdc27 in Figure 7F), it is possible that the formation of long flaps is reduced. Under these circumstances, Cdc24–Dna2 activity becomes dispensable and the step illustrated in Figure 7C is bypassed: in this way, cdc27-D1 and cdc27-D3 are able to suppress cdc24-M38 and dna2-C2 at their restrictive temperatures. The shorter flaps generated under these conditions (e.g. in cdc27-D1 cdc24-M38 cells) are then presumably processed by Fen1. When Cdc24 function is partially impaired, as in cdc24-M38 cells grown at their permissive temperature, lengthy uncleaved flap structures may transiently accumulate (Figure 7G). This appears to lead to the formation of slippage structures between short direct repeats (Figure 7H) and the generation of sequence duplications. Confirmation of this model will require further biochemical investigation.
Figure 7.
Model for Okazaki fragment processing in fission yeast. Left, schematic showing Cdc24-Dna2 processing of 5′ flap structures, from the first approach of the Pol δ-PCNA complex (A), through displacement synthesis (B) and Cdc24-Dna2 binding (C; for clarity, RPA is omitted), to Fen1 cleavage (D) and Cdc17 catalysed ligation (E). Right, upper part: when Pol δ processivity is reduced as a result of impairment of the Cdc27-PCNA interaction, a short flap only is generated (F) that can be processed by Fen1(D). Right, lower part: when Cdc24-Dna2 function is impaired, transient stabilisation of long flaps (G) can result in repeat slippage. See text for further discussion.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank our colleagues and friends in Edinburgh, Daejeon and elsewhere for their assistance with this work, in particular M. Yamamoto (Kazusa DNA Research Institute) for the pol3+ gene and cDNA, K. Okazaki (University of Tokyo) for S.pombe genomic DNA libraries, and J. Bähler (Sanger Institute, Hinxton, UK) for pFA6a-13Myc-kanMX6. H.T. was supported by a JSPS Postdoctoral Fellowship for Research Abroad and by the Wellcome Trust. S.A.M. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science.
REFERENCES
- 1.Hübscher U. and Seo,Y.S. (2001) Replication of the lagging strand: a concert of at least 23 polypeptides. Mol. Cells, 12, 149–157. [PubMed] [Google Scholar]
- 2.Zuo S., Gibbs,E., Kelman,Z., Wang,T.S., O'Donnell,M., MacNeill,S.A. and Hurwitz,J. (1997) DNA polymerase δ isolated from Schizosaccharomyces pombe contains five subunits. Proc. Natl Acad. Sci. USA, 94, 11244–11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reynolds N., Watt,A., Fantes,P.A. and MacNeill,S.A. (1998) Cdm1, the smallest subunit of DNA polymerase δ in the fission yeast Schizosaccharomyces pombe, is non-essential for growth and division. Curr. Genet., 34, 250–258. [DOI] [PubMed] [Google Scholar]
- 4.Podust V.N., Chang,L.S., Ott,R., Dianov,G.L. and Fanning,E. (2002) Reconstitution of human DNA polymerase δ using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J. Biol. Chem., 277, 3894–3901. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez Garcia J., Ciufo,L.F., Yang,X., Kearsey,S.E. and MacNeill,S.A. (2004) The C-terminal zinc finger of the catalytic subunit of DNA polymerase δ is responsible for direct interaction with the B-subunit. Nucleic Acids Res., 32, 3005–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacNeill S.A., Moreno,S., Reynolds,N., Nurse,P. and Fantes,P.A. (1996) The fission yeast Cdc1 protein, a homolog of the small subunit of DNA polymerase δ, binds to Pol3 and Cdc27. EMBO J., 15, 4613–4628. [PMC free article] [PubMed] [Google Scholar]
- 7.Warbrick E. (1998) PCNA binding through a conserved motif. Bioessays, 20, 195–199. [DOI] [PubMed] [Google Scholar]
- 8.Huang M.E., LeDouarin,B., Henry,C. and Galibert,F. (1999) The Saccharomyces cerevisiae protein YJR043C (Pol32) interacts with the catalytic subunit of DNA, polymerase α and is required for cell cycle progression in G2/M. Mol. Gen. Genet., 260, 541–550. [DOI] [PubMed] [Google Scholar]
- 9.Johansson E., Garg,P. and Burgers,P.M. (2004) The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem., 279, 1907–1915. [DOI] [PubMed] [Google Scholar]
- 10.Gray F.C., Pöhler,J.R.C., Warbrick,E. and MacNeill,S.A. (2004) Mapping and mutation of the conserved DNA polymerase interaction motif (DPIM) located in the C-terminal domain of fission yeast DNA polymerase δ subunit Cdc27. BMC Mol. Biol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gould K.L., Burns,C.G., Feoktistova,A., Hu,C.P., Pasion,S.G. and Forsburg,S.L. (1998) Fission yeast cdc24+ encodes a novel replication factor required for chromosome integrity. Genetics, 149, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka H., Tanaka,K., Murakami,H. and Okayama,H. (1999) Fission yeast Cdc24 is a replication factor C- and proliferating cell nuclear antigen-interacting factor essential for S-phase completion. Mol. Cell Biol., 19, 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang H.Y., Choi,E., Bae,S.H., Lee,K.H., Gim,B.S., Kim,H.D., Park,C., MacNeill,S.A. and Seo,Y.S. (2000) Genetic analyses of Schizosaccharomyces pombe dna2+ reveal that dna2 plays an essential role in Okazaki fragment metabolism. Genetics, 155, 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 15.Rothstein R.J. (1983) One-step gene disruption in yeast. Methods Enzymol, 101, 202–211. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka H., Ryu,G.H., Seo,Y.S., Tanaka,K., Okayama,H., MacNeill,S.A. and Yuasa,Y. (2002) The fission yeast pfh1+ gene encodes an essential 5′ to 3′ DNA helicase required for the completion of S-phase. Nucleic Acids Res., 30, 4728–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryu G.H., Tanaka,H., Kim,D.H., Kim,J.H., Bae,S.H., Kwon,Y.N., Rhee,J.S., MacNeill,S.A. and Seo,Y.S. (2004) Genetic and biochemical analyses of Pfh1 DNA helicase function in fission yeast. Nucleic Acids Res., 32, 4205–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inge-Vechtomov S., Zhouravleva,G. and Philippe,M. (2003) Eukaryotic release factors (eRFs) history. Biol. Cell, 95, 195–209. [DOI] [PubMed] [Google Scholar]
- 19.Nasmyth K. and Nurse,P. (1981) Cell division cycle mutants altered in dna-replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 182, 119–124. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds N., Warbrick,E., Fantes,P.A. and MacNeill,S.A. (2000) Essential interaction between the fission yeast DNA polymerase δ subunit Cdc27 and Pcn1 (PCNA) mediated through a C-terminal p21Cip1-like PCNA binding motif. EMBO J., 19, 1108–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuo S., Bermudez,V., Zhang,G., Kelman,Z. and Hurwitz,J. (2000) Structure and activity associated with multiple forms of Schizosaccharomyces pombe DNA polymerase δ. J. Biol. Chem., 275, 5153–5162. [DOI] [PubMed] [Google Scholar]
- 22.Liu V.F., Bhaumik,D. and Wang,T.S. (1999) Mutator phenotype induced by aberrant replication. Mol. Cell Biol., 19, 1126–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fantes P.A. and Creanor,J. (1984) Canavanine resistance and the mechanism of arginine uptake in the fission yeast Schizosaccharomyces pombe. J. Gen. Micro., 130, 3265–3273. [DOI] [PubMed] [Google Scholar]
- 24.Sychrova H., Chevallier,M.R., Horak,J. and Kotyk,A. (1992) Thialysine-resistant mutants and uptake of lysine in Schizosaccharomyces pombe. Curr. Genet., 21, 351–355. [DOI] [PubMed] [Google Scholar]
- 25.Budd M.E., Choe,W.C. and Campbell,J.L. (1995) Dna2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J. Biol. Chem., 270, 26766–26769. [DOI] [PubMed] [Google Scholar]
- 26.Budd M.E. and Campbell,J.L. (1995) Dna2, a new yeast gene required for dna-replication encodes a DNA helicase. J. Cell Biochem., 306–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budd M.E. and Campbell,J.L. (1995) A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl Acad. Sci. USA, 92, 7642–7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Budd M.E. and Campbell,J.L. (1997) A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell Biol., 17, 2136–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiorentino D.F. and Crabtree,G.R. (1997) Characterization of Saccharomyces cerevisiae dna2 mutants suggests a role for the helicase late in S phase. Mol. Biol. Cell., 8, 2519–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bae S.H., Choi,E., Lee,K.H., Park,J.S., Lee,S.H. and Seo,Y.S. (1998) Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem., 273, 26880–26890. [DOI] [PubMed] [Google Scholar]
- 31.Bae S.H. and Seo,Y.S. (2000) Characterization of the enzymatic properties of the yeast Dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem., 275, 38022–38031. [DOI] [PubMed] [Google Scholar]
- 32.Bae S.H., Bae,K.H., Kim,J.A. and Seo,Y.S. (2001) RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature, 412, 456–461. [DOI] [PubMed] [Google Scholar]
- 33.Bae S.H., Kim,J.A., Choi,E., Lee,K.H., Kang,H.Y., Kim,H.D., Kim,J.H., Bae,K.H., Cho,Y., Park,C. et al. (2001) Tripartite structure of Saccharomyces cerevisiae Dna2 helicase/endonuclease. Nucleic Acids Res., 29, 3069–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tishkoff D.X., Filosi,N., Gaida,G.M. and Kolodner,R.D. (1997) A novel mutation avoidance mechanism dependent on S.cerevisiae RAD27 is distinct from DNA mismatch repair. Cell, 88, 253–263. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y., Kao,H.I. and Bambara,R.A. (2004) Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem., 73, 589–615. [DOI] [PubMed] [Google Scholar]
- 36.Bermudez V.P., MacNeill,S.A., Tappin,I. and Hurwitz,J. (2002) The influence of the Cdc27 subunit on the properties of the Schizosaccharomyces pombe DNA polymerase δ. J. Biol. Chem., 277, 36853–36862. [DOI] [PubMed] [Google Scholar]