ABSTRACT
PhoU, a conserved protein that has been proposed to coordinate phosphate import, is a negative regulator of drug tolerance in most bacteria. In Staphylococcus epidermidis, the role of PhoU in biofilm formation and drug tolerance has not yet been investigated. Two PhoU homologs in the genome of S. epidermidis have been identified by the presence of the conserved motif E(D)XXXD of PhoU. We separately constructed ΔphoU1 and ΔphoU2 mutants of S. epidermidis strain 1457. The ΔphoU2 mutant displayed growth retardation, a weakened biofilm formation capacity, a higher sensitivity to H2O2, and reduced tolerance to multiple antibiotics. However, deletion of phoU1 had no effect on those. We compared the transcriptome profiles of the ΔphoU2 and ΔphoU1 mutants with that of the parent strain. In the ΔphoU2 mutant, expression of genes related to inorganic phosphate uptake was significantly upregulated (pst operon) and the levels of intracellular inorganic polyphosphate (polyP) were increased. In the ΔphoU2 mutant, expression of enzymes in the pentose phosphate pathway (PPP) was downregulated and less NADP (NADPH) was detected, consistent with the high sensitivity to H2O2 and the growth retardation of the ΔphoU2 mutant. The upregulated expression of ATP synthase was consistent with the high intracellular ATP content in the ΔphoU2 mutant, which may have been related to the lower drug tolerance of the ΔphoU2 mutant. This study demonstrates that PhoU2, but not PhoU1, in S. epidermidis regulates bacterial growth, biofilm formation, oxidative stress, and drug tolerance in association with alterations to inorganic phosphate metabolism, the pentose phosphate pathway, galactose metabolism, the tricarboxylic acid (TCA) or citric cycle, glycolysis and gluconeogenesis, and respiratory reactions.
IMPORTANCE PhoU is widely conserved throughout the bacterial kingdom and plays an important role in response to stress and metabolic maintenance. In our study, two PhoU homologs were found in S. epidermidis. The function of phoU2, but not phoU1, in S. epidermidis is related to growth, drug tolerance, the oxidative stress response, polyP levels, and ATP accumulation. In addition, phoU2 regulates biofilm formation. Hence, phoU2 is a regulator of both drug tolerance and biofilm formation, which are two bacterial properties that present major challenges to the clinical treatment of infections. Analysis of differential gene expression revealed that phoU2 is involved in fundamental metabolic processes, such as the PPP pathway. These findings indicate that phoU2 is a crucial regulator in S. epidermidis.
KEYWORDS: biofilm, Staphylococcus epidermidis, tolerance
INTRODUCTION
Staphylococcus epidermidis is a common opportunistic pathogen that is present on human skin and mucosal surfaces. The pathogenicity of S. epidermidis is mainly due to biofilm formation on foreign devices, such as catheters, heart valves, and prostheses (1, 2). The bacteria within the biofilm are protected against killing by antibiotics and the host immune system, contributing to the increasing emergence of resistance to antimicrobial drugs and to the establishment of persistent infections (3, 4). PhoU, the phosphate transport system regulatory protein, is now known to be a negative regulator of drug tolerance in Escherichia coli, Mycobacterium tuberculosis, and Pseudomonas aeruginosa (5–8), whereas the possible role of PhoU in biofilm formation and drug tolerance in S. epidermidis has not been investigated.
On the basis of a crystal structure analysis of PhoU in Thermotoga maritima (9), multinuclear iron clusters [E(D)XXXD] have been identified to be the conserved motif of the protein. According to the conserved motif, one homolog of phoU is found in E. coli, P. aeruginosa, and Streptococcus, while two phoU homologs are found in T. maritima, M. tuberculosis, Mycobacterium marinum, and Staphylococcus aureus. In E. coli and P. aeruginosa, phoU is located in the pst operon, which contains four other genes: pstS, pstA, pstC, and pstB. PstS is a periplasmic inorganic phosphate (Pi)-binding protein that captures and transfers Pi to the channel formed by the integral proteins PstC and PstA in the cytoplasmic membrane. PstB is an ATPase that provides the energy for transport (10). The last gene in the operon, phoU, encodes a protein that does not have a defined function. In 2007, it was reported that phoU in E. coli plays an important role in the development of multidrug-tolerant bacteria (5). In P. aeruginosa, phoU is a negative regulator of intracellular ppGpp and polyphosphate (polyP), but it does not regulate biofilm formation (8). In M. tuberculosis and M. marinum, there are two phoU homologs in the genome, phoY1 and phoY2. However, neither phoY1 nor phoY2 is located in the pst operon. In the two mycobacterial species, phoY2 was shown to be a functional homolog of phoU, regulating the generation of multidrug-tolerant bacteria and maintaining metabolic homeostasis and adaptation to stress conditions (6, 7). In the S. aureus (NCTC8325) genome, there are two phoU homologs (SAOUHSC_01384 and SAOUHSC_00669). In S. aureus, SAOUHSC_01384 resides in the pst operon as phoU, while SAOUHSC_00669 (pitR) also contains the conserved motif of phoU and is located upstream of pitA (a phosphate uptake gene). A point mutation in pitA in S. aureus resulted in bacteria that were sensitized to daptomycin, and SAOUHSC_00669 was found to be required for daptomycin sensitivity (11, 12). In the present study, protein motif analysis of S. epidermidis ATCC 35984 revealed two genes that were homologous to phoU (serp0956 and serp0316). serp0956, found in the pst operon, was named phoU1, while serp0316, located upstream of a hypothetical protein with a high degree of homology to pitR of S. aureus, was named phoU2.
In staphylococci, biofilm formation and drug tolerance are regulated by multiple regulatory factors (13, 14). In this study, we focused on investigating the roles of PhoU homologs in the biofilm formation and drug tolerance of S. epidermidis. In S. epidermidis strain 1457 (SE1457), either phoU1 or phoU2 was deleted by allelic replacement to create the ΔphoU1 and ΔphoU2 mutants, respectively. The effects of these deletions on bacterial growth, biofilm formation, drug tolerance, and oxidative stress were investigated. Comparison of the transcriptomes of the ΔphoU2 mutant and the parent strain revealed the differentially expressed genes (DEGs) involved in the pentose phosphate (PPP) pathway, galactose metabolism, the trichloroacetic acid (TCA) cycle, glycolysis and gluconeogenesis, and respiratory chain reactions. The involvement of some of the DEGs in metabolic processes, such as polyP accumulation, ATP accumulation, and the pentose phosphate pathway, was validated. We conclude that phoU2 probably regulates bacterial growth, biofilm formation, oxidative stress responses, and drug tolerance in S. epidermidis. In contrast, phoU1 has no obvious effect on biological activity in S. epidermidis.
RESULTS
The two phoU loci identified in the genome of S. epidermidis.
In the genome of the S. epidermidis ATCC 35984 strain (GenBank accession number CP000029), two PhoU gene homologs, serp0956 and serp0316, were identified by bioinformatics analysis (https://www.ncbi.nlm.nih.gov/genome/?term=RP62a) on the basis of the conserved motif E(D)XXXD of Thermotoga maritima. serp0956 is located in the pst operon, similarly to the phoU gene in E. coli, which is denoted as encoding a phosphate transport system regulatory protein and designated phoU1 in this study. serp0316, denoted phoU2, encodes a hypothetical protein, is located far from the pst operon, and was cotranscribed with the adjacent gene, serp0317 (Fig. 1). The two PhoU homologs in S. epidermidis shared 12.5% identity and 30.1% consensus sequences.
FIG 1.
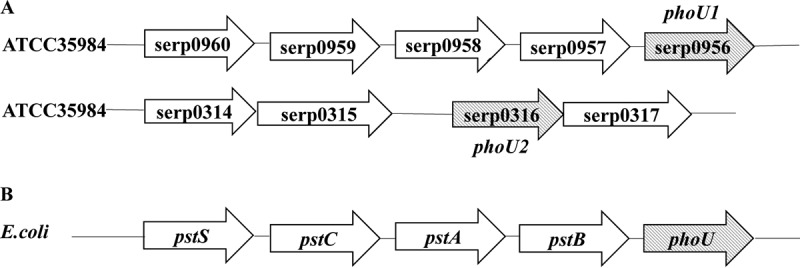
Two genes homologous to phoU, phoU1 and phoU2, were found in S. epidermidis RP62A by motif analysis according to the PhoU conserved motifs [E(D)XXXD] of Thermotoga maritima. (A) Genetic locations of phoU1 and phoU2 in S. epidermidis RP62A. (B) Genetic organization of the pst operon and phoU in E. coli.
Construction of phoU1 and phoU2 deletion strains.
To identify the function of phoU1 or phoU2 in S. epidermidis, mutants of the SE1457 strain with a deletion of phoU1 or phoU2 were constructed using the temperature-sensitive plasmid pKOR1. The deletion mutants were verified by PCR, reverse transcription (RT)-quantitative PCT (qPCR), and direct sequencing and are referred to as the ΔphoU1 and ΔphoU2 mutants, respectively. The complemented ΔphoU2 strain was constructed using shuttle vector pCN51 and named SE1457 ΔphoU2/pCN51::phoU2. The ΔphoU2 strain containing the empty vector pCN51 was designated SE1457 ΔphoU2/pCN51.
Growth curves of the ΔphoU1 and ΔphoU2 mutants.
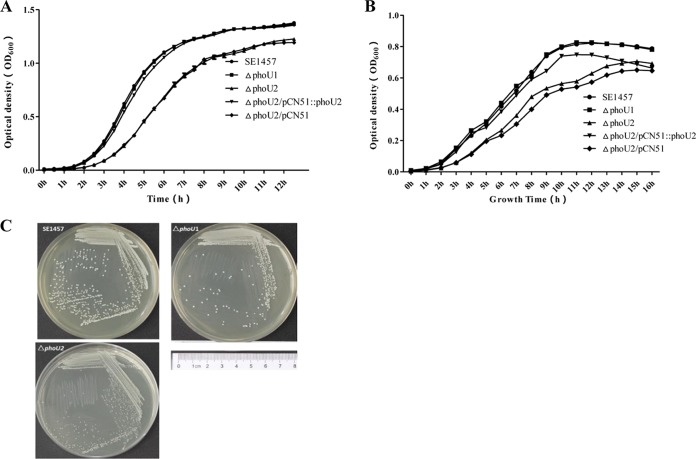
To evaluate the effect of phoU1 or phoU2 knockout on the growth of S. epidermidis, growth curves of the ΔphoU1 and ΔphoU2 mutants and the parent strain (SE1457) were generated under oxic and microaerobic conditions.
Under oxic conditions, in comparison to the parent strain, the ΔphoU2 mutant displayed a marked reduction in growth (Fig. 2A). However, the ΔphoU1 mutant displayed growth curves and colony sizes similar to those of the parent strain when the strains were incubated under the same conditions. By culture in liquid tryptic soya broth (TSB) medium at 37°C for 6 h, the optical density at 600 nm (OD600) of the ΔphoU2 mutant reached 1.26 ± 0.343, and that of the parent strain was 2.41 ± 0.078. On solid culture medium (TSB agar), the colony size of the ΔphoU2 mutant was much smaller than that of the parent strain (Fig. 2C). The expression of phoU2 by the pCN51 vector restored the growth of the ΔphoU2 mutant to the level of the parent strain.
FIG 2.
Effect of phoU2 deletion on the growth of S. epidermidis. Overnight cultures of the ΔphoU1, ΔphoU2, and SE1457 strains were diluted 1:200 into 10 ml TSB in a conical flask in a volume of 100 ml and incubated with shaking at 220 rpm. Bacterial growth was monitored by measuring the OD600 for 12 h. Data (means ± SDs) are from three independent experiments. (A) Growth curves of the ΔphoU1 and ΔphoU2 mutants under oxic conditions. (B) Growth curves of ΔphoU1 and ΔphoU2 mutants under microaerobic conditions. (C) Representative images showing the colony morphology of the ΔphoU1 and ΔphoU2 mutants and the SE1457 parent strain grown on TSB agar plates at 37°C for 24 h under oxic conditions. The results represent those from one of three independent experiments.
Under microaerobic conditions, the growth rates of the ΔphoU1 and ΔphoU2 mutants and SE1457 in liquid medium were measured by recording the OD600 of the cultures. After 6 h of incubation, the OD600 of the ΔphoU2 mutant was 0.265 ± 0.04, which was much less that than that of SE1457 (0.418 ± 0.03) (Fig. 2B). However, no differences in the growth curve could be detected in the ΔphoU1 mutant when the strains were incubated under the same conditions.
Morphology and autolysis of the ΔphoU1 and ΔphoU2 mutants.
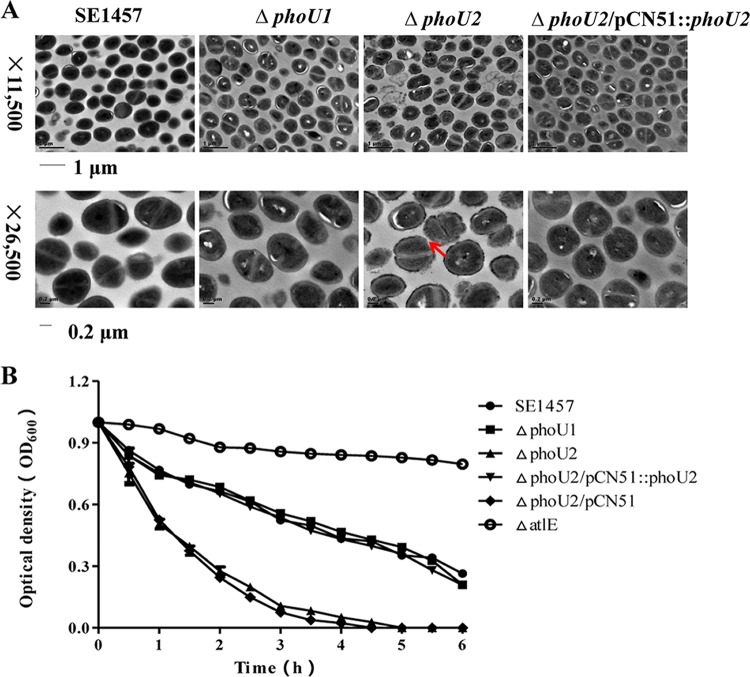
The morphologies of the ΔphoU1 and ΔphoU2 mutants and the parent strain were observed using transmission electron microscopy (TEM). A rough cell wall and thin cytoplasm were observed in the ΔphoU2 mutant (magnification, ×26,500), while the morphology of the ΔphoU1 mutant was similar to that of the parent strain (Fig. 3A).
FIG 3.
(A) Bacterial morphology of the ΔphoU1 and ΔphoU2 mutants observed by TEM. The ultrastructure of the log-phase bacteria was observed by transmission electron microscopy (Philips Tecnai-12 Biotwin). Arrow, disruption of the cell wall in the ΔphoU2 mutant. (B) Autolysis of the ΔphoU1 and ΔphoU2 mutants induced by Triton X-100. Overnight cultures were suspended in Triton X-100 autolysis buffer (50 mM glycine, pH 8.0, containing 0.01% Triton X-100) to an initial OD600 of approximately 1.0, and the rates of autolysis were monitored on the basis of the decrease in the OD600 value over time.
The autolysis capacity of the ΔphoU1 and ΔphoU2 mutants and the parent strain was then assessed by Triton X-100 induction. Mid-exponential-phase cultures (6 h) of the ΔphoU1 and ΔphoU2 mutants and SE1457 were adjusted to an OD600 of 1.0 and incubated with 0.05% Triton X-100 for 5 h, and the autolysis rate was measured by determination of the OD600 using a spectrophotometer. After Triton X-100 induction, the autolysis rate of the ΔphoU2 mutant reached 88% after treatment for 3 h, which was significantly higher than that of SE1457 (46%). The autolysis rate of ΔphoU1 (43%) was similar to that of the parent strain (Fig. 3B).
Biofilm formation of the ΔphoU1 and ΔphoU2 mutants under static or hydrodynamic conditions.
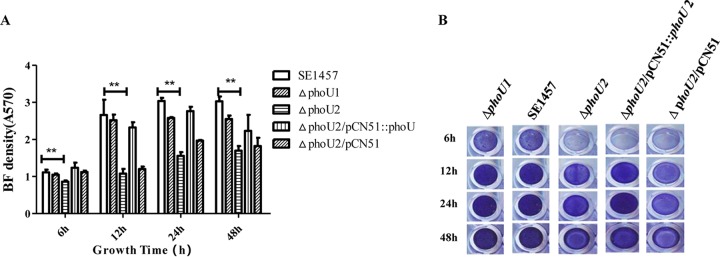
Under static conditions, the polystyrene microtiter plate assay and the confocal laser scanning microscopy (CLSM) observation assay were performed to evaluate the role of phoU1 or phoU2 in biofilm formation. Bacterial biofilm formation was monitored at 6, 12, 24, and 48 h on microtiter plates stained with crystal violet, and the OD570 was read. The mature biofilm of the ΔphoU2 mutant (OD570, 1.56 ± 0.10) was significantly decreased compared with that of the parent strain (OD570, 3.04 ± 0.08) after incubation for 24 h, and the ΔphoU1 deletion (OD570, 2.58 ± 0.02) had little effect on biofilm formation (Fig. 4). The complemented ΔphoU2/pCN51::phoU2 strain (OD570, 2.77 ± 0.12) showed a restored biofilm formation ability.
FIG 4.
Biofilm (BF) formation by the ΔphoU1 and ΔphoU2 mutants on microtiter plates. Overnight cultures of the S. epidermidis strains were diluted 1:200 with fresh TSB, added to 96-well polystyrene plates in triplicate, and cultured under static conditions for 6 h, 12 h, 24 h, and 48 h. After the biofilms were washed, they were stained with crystal violet. The OD570s of the plates were analyzed. The experiments were repeated three times, and the data represent means ± SDs. **, P < 0.01.
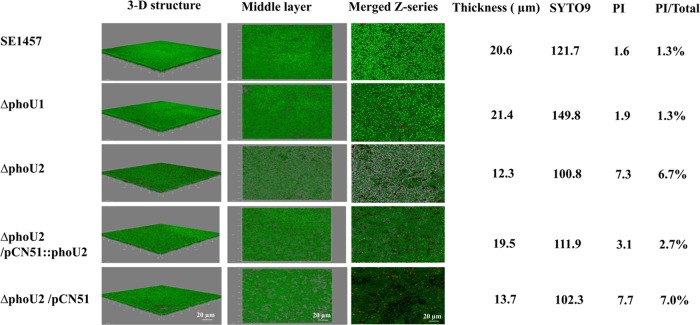
Furthermore, the biofilm formation ability of the ΔphoU2 mutant in vitro was examined by confocal laser scanning microscopy. After incubation at 37°C for 24 h, SE1457 formed a compact, thick biofilm on a glass coverslip in a cell culture dish. In contrast, the biofilm of the ΔphoU2 mutant was much thinner than that of the parent strain. Additionally, the dead cell ratio in the biofilm of the ΔphoU2 mutant (6.7%) was 5-fold higher than that in the biofilm of the parent strain (1.3%) (Fig. 5).
FIG 5.
Biofilms of the ΔphoU1 and ΔphoU2 mutants observed by CLSM. Twenty-four-hour-old biofilms of SE1457 and the ΔphoU1 and ΔphoU2 mutants were grown on a cover glass in a cell culture dish and observed by CLSM. Three-dimensional (3-D) structural images (zoom 1, ×63 magnification) were reconstructed, and the thickness of the biofilms was measured using Imaris software. Viable and dead cells were stained green (SYTO9) and red (PI), respectively. The amount of fluorescence in the middle layer of the biofilm was determined using ImageJ software (zoom 3, ×63 magnification). The PI/total florescence value indicates the proportion of dead cells within the biofilm. The images and values are representative of those from one of three independent experiments.
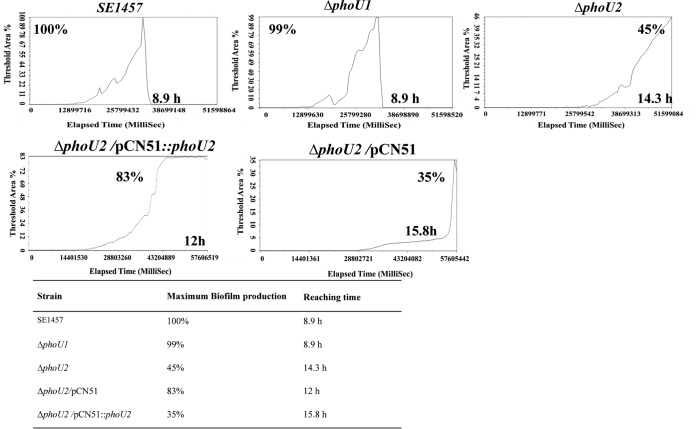
Under hydrodynamic conditions, the biofilm formation of the ΔphoU2 mutant strain was analyzed using a BioFlux 1000 device with a flow speed of 0.15 dyne/cm2. The maximum size of the biofilm of the ΔphoU2 mutant observed in the channel was 2-fold smaller than that of the biofilm of the parent strain and the complemented strain, and the time required to form the maximum biofilm was 5.4 h longer for the ΔphoU2 mutant strain than for the parent strain (Fig. 6). The biofilm formation of the ΔphoU1 mutant in the hydrodynamic state appeared to be similar to that of the parent strain. The complemented ΔphoU2/pCN51::phoU2 strain could partially restore the biofilm formation capacity under hydrodynamic conditions.
FIG 6.
Biofilm formation by the ΔphoU1 and ΔphoU2 mutants under hydrodynamic conditions. Overnight cultures of the S. epidermidis strains were diluted 1:200 with fresh TSB and added to BioFlux 48-well plates. The bacteria were then cultured under hydrodynamic conditions with a shear setting of 0.15 dyne/cm2. A BioFlux 1000 system (Fluxion Biosciences) with a Leica microscope and temperature-controlled housing was used for all imaging experiments. Automated microscopy and image processing were performed with BioFlux Montage software. Images were automatically acquired every 10 min at multiple stage positions with bright-field illumination; images were also acquired in the red channel using a 200-ms exposure time. The background-corrected average pixel intensity per image was used to quantify the biofilm formation by the different strains. The curve was generated on the basis of the images. A synthesis of the images is shown in Movies S1 to S5 in the supplemental material. Each figure represents the results of one of three independent experiments.
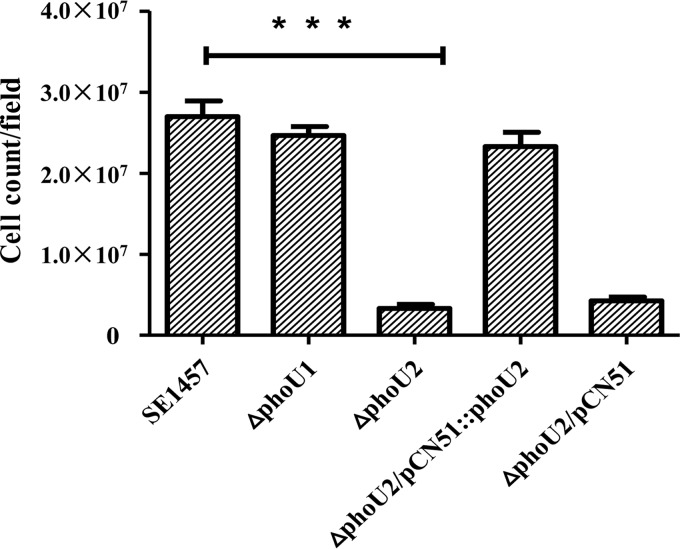
The initial attachment phase is characterized as the first phase of the process of Staphylococcus epidermidis biofilm formation. The initial attachment capacity of the ΔphoU2 mutant was determined, and the attached cells were counted using ImageJ software. There were fewer attached cells for the ΔphoU2 mutant (3.32 × 106) than for the parent strain (2.70 × 107) and the complemented strain ΔphoU2/pCN51::phoU2 (2.33 × 107). The number of attached cells of the ΔphoU1 mutant (2.47 × 107) was similar to the number of attached cells of the parent strain (Fig. 7).
FIG 7.
Initial attachment of the ΔphoU1 and ΔphoU2 mutants on polystyrene plates. Log-phase bacterial cultures in TSB were adjusted to an OD600 of 0.1 with PBS, and 5-ml aliquots were added to a 6-well polystyrene plate. After incubation at 37°C for 30 min, each well was washed three times with PBS, and the adhered cells were observed and photographed. The amounts of attached bacterial cells of the ΔphoU1 and ΔphoU2 mutants and SE1457 are indicated. The results (means ± SDs) are from three independent experiments. ***, P < 0.001.
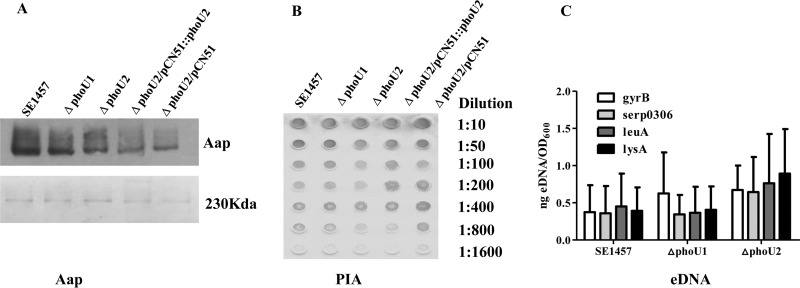
To investigate the effect of the phoU2 deletion on biofilm matrix production, the release of accumulation-associated protein (Aap), polysaccharide intercellular adhesion (PIA), and extracellular DNA (eDNA) was determined for SE1457 and the ΔphoU1 and ΔphoU2 mutants. The production of Aap was reduced in the biofilm of the ΔphoU2 mutant compared with that in the biofilm of the parent strain (Fig. 8A), as determined by Western blotting with monoclonal antibody 18B6 (MAb18B6) against the Aap protein B repeat region. PIA production was similar in the ΔphoU2 mutant biofilm and the parent strain, as determined semiquantitatively with a wheat germ agglutinin (WGA)-horseradish peroxidase (HRP) conjugate using a dot blot 96 system (Fig. 8B). The relative concentration of eDNA in the 24-h-old biofilm of the ΔphoU2 mutant was similar to that in the parent strain (Fig. 8C). All P values were >0.05, so there were no differences in the levels of transcription of serp0306, leuA, or lysA between biofilm bacteria of the PhoU mutants and the parent strain. The levels of PIA, Aap, and eDNA production were similar in the ΔphoU1 mutant and the parent strain.
FIG 8.
Effects of the ΔphoU1 and ΔphoU2 mutants on extracellular matrix biosynthesis by S. epidermidis. (A) Aap expression in the ΔphoU1 and ΔphoU2 mutants. Twenty-four-hour-old biofilms and 12-h-old planktonic bacteria were collected after they were washed with PBS. Lysostaphin-treated samples with identical OD600s were centrifuged at 20,000 × g for 30 min at 4°C. The supernatant was separated by 7% SDS-PAGE, and the gel pieces containing Aaps were used for Western blotting (top).The remaining gel pieces were stained with Coomassie blue as an endogenous reference (bottom). MAb18B6 (10 ng/ml) was used as the primary antibody. Immunoreactivity was detected using an ECL Western blotting system after incubation with HRP-conjugated secondary antibody. (B) PIA biosynthesis was semiquantified using a dot blot assay with WGA. Twenty-four-hour-old biofilms were scraped off and suspended in EDTA. Serial dilutions of the PIA assay extracts were spotted onto nitrocellulose membranes, subsequently incubated with WGA conjugated to HRP, and visualized by chromogenic detection. (C) eDNA quantified by qPCR of four chromosomal loci (gyrB, serp0306, leuA, and lysA). The OD600s of unwashed 24-h-old biofilms were measured for normalization to the biofilm biomass, and then the biofilms were used for eDNA isolation by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation. The results are presented as the amount of eDNA per biofilm biomass (means ± SDs) from three independent experiments.
Antibiotic tolerance of the ΔphoU1 and ΔphoU2 mutants.
Antibiotic-tolerant bacteria were identified using a modification of a procedure described by Li and Zhang (5). Both the MIC and the minimal bactericidal concentration (MBC) for the ΔphoU1 and ΔphoU2 mutants were similar to those for the parent strain. To determine the antibiotic concentrations that ensured that only drug-tolerant bacterial cells survived, killing curves were determined for each antibiotic used in the present study. To detect the drug tolerance of the bacterial mutants, in which the size of the population of bacteria was not decreased with an increase in the antibiotic concentration, the concentrations of antibiotics were as follows: vancomycin, 75 μg/ml (96× MIC); levofloxacin, 75 μg/ml (128× MIC); amikacin, 50 μg/ml (128× MIC).
An overnight culture (16 h) of S. epidermidis was inoculated into fresh TSB (at 1:100) containing the antibiotics at the specific concentrations, and the culture was incubated at 37°C for 5 days. At different time points, the surviving bacteria were counted. After exposure to the three antibiotics for 72 h, the antibiotic tolerance of the ΔphoU2 mutant was dramatically reduced, and no viable bacteria were detected. In contrast, the ΔphoU1 mutant displayed an antibiotic tolerance similar to that of the parent strain (greater than 104 CFU) (Table 1). The complementation of the ΔphoU2 mutant strain restored the antibiotic tolerance.
TABLE 1.
Survival of the ΔphoU1 and ΔphoU2 mutants and the parent strain with antibiotic exposure over timea
| Time point | No. of CFU/ml |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SE1457 |
ΔphoU1 mutant |
ΔphoU2 mutant |
ΔphoU2/pCN51::phoU2 strain |
ΔphoU2/pCN51 strain |
|||||||||||
| Van | Lev | Ami | Van | Lev | Ami | Van | Lev | Ami | Van | Lev | Ami | Van | Lev | Ami | |
| Start | 8 × 109 | 8 × 109 | 8 × 109 | 8.7 × 109 | 8.7 × 109 | 8.7 × 109 | 9 × 109 | 9 × 109 | 9 × 109 | 8.5 × 109 | 8.5 × 109 | 8.5 × 109 | 8.7 × 109 | 8.7 × 109 | 8.7 × 109 |
| 12 h | 6 × 106 | 4 × 106 | 7.2 × 106 | 8 × 106 | 7.8 × 106 | 4.4 × 106 | 8 × 105 | 5 × 104 | 6 × 105 | 6.8 × 106 | 5 × 106 | 7.8 × 106 | 7.6 × 106 | 5.7 × 104 | 4 × 105 |
| 24 h | 3.2 × 106 | 3.6 × 106 | 5 × 105 | 6 × 106 | 5 × 106 | 4 × 106 | 5 × 105 | 2 × 103 | 1 × 105 | 4 × 106 | 3.5 × 106 | 2 × 106 | 3.5 × 105 | 5 × 103 | 2.4 × 104 |
| 36 h | 9.2 × 105 | 7 × 105 | 4 × 105 | 1.2 × 106 | 2.4 × 106 | 2 × 106 | 5.4 × 105 | 0 | 6 × 104 | 2.2 × 106 | 8 × 105 | 7 × 105 | 4 × 104 | 0 | 2 × 103 |
| 48 h | 1.3 × 106 | 5 × 105 | 8.8 × 104 | 1.28 × 106 | 3 × 106 | 5 × 105 | 4 × 104 | 0 | 1.2 × 104 | 7 × 105 | 8 × 105 | 4.5 × 105 | 2.4 × 103 | 0 | 0 |
| 72 h | 4.4 × 106 | 2 × 105 | 6 × 104 | 2.57 × 106 | 5 × 105 | 4 × 105 | 0 | 0 | 0 | 5 × 105 | 4.6 × 105 | 9 × 104 | 0 | 0 | 0 |
| 96 h | 6 × 106 | 7 × 105 | 4 × 104 | 9 × 106 | 6.6 × 105 | 1 × 105 | 0 | 0 | 0 | 7.2 × 105 | 5 × 105 | 7.5 × 104 | 0 | 0 | 0 |
| 120 h | 1.9 × 105 | 6 × 105 | 1.9 × 104 | 6 × 106 | 4 × 105 | 8 × 104 | 0 | 0 | 0 | 2.4 × 105 | 4.5 × 105 | 5 × 104 | 0 | 0 | 0 |
The susceptibilities of stationary-phase cultures of the ΔphoU1 and ΔphoU2 mutants and the SE1457 parent strain to vancomycin (Van; 75 μg/ml), levofloxacin (Lev; 75 μg/ml), and amikacin (Ami; 50 μg/ml) were determined. The numbers of CFU were determined at different times of exposure of stationary-phase cultures of these strains to the indicated antibiotics.
Comparison of the transcriptomes of the ΔphoU2 mutant and the parent strain.
To compare the transcriptional profile of the ΔphoU1 or ΔphoU2 mutant with that of SE1457, RNA was extracted from logarithmic-phase (6 h) and stationary-phase (10 h) bacteria and detected by transcriptome sequencing (RNA-Seq). The sequencing libraries were prepared in triplicate for the ΔphoU1 and ΔphoU2 mutants and the parent strain. For each biological replicate, 10 million raw reads were generated. After the removal of ambiguous and low-quality reads, more than 90% of the reads mapped to strain ATCC 35984.
A gene with a false discovery rate (FDR)-adjusted P value of less than 0.05 (t test), a q value of less than 0.05, and at least a 1.5-fold change in the transcript level between the mutant and the parent strain was considered to be differentially expressed. In logarithmic phase (6 h), 945 genes were identified to be differentially expressed between the ΔphoU2 mutant and the parent strain; among these, 474 genes were upregulated and 471 were downregulated. In the stationary phase (10 h), 995 DEGs were identified; among these, 716 genes were upregulated and 279 were downregulated. By comparison and analysis, 439 DEGs were identified in the different phases of both the ΔphoU2 mutant and the parent strain. Among these genes, 256 showed the same expression tendency. However, there were only 92 genes differentially expressed between the ΔphoU1 mutant and the parent strain during logarithmic phase and 2 DEGs during the stationary phase. Therefore, we focused on analyzing the genes differentially expressed between the ΔphoU2 mutant and the parent strain. We selected 70 of the DEGs for validation by RT-qPCR. Among them, the transcription of 62 genes in the ΔphoU2 mutant was consistent with the tendency observed by RNA-Seq. The other 8 genes determined by RT-qPCR displayed a fold change in expression below the cutoff value of 2.
Among the DEGs, the transcription of yycFG, an essential two-component system for regulating bacterial growth, in the ΔphoU2 mutant was downregulated 3-fold compared with its expression in the parent strain (15). Transcription of the anaerobic growth-regulated genes pflA, nrdD, and nrdG was downregulated in the ΔphoU2 mutant (16–19). The expression of rsbU (a sigma factor B regulatory protein), which is involved in biofilm formation, was also downregulated (20). Transcription of the autolysis genes ssaA (serp2136, serp1880, serp1884, serp2120) and serp0318 was upregulated in the ΔphoU2 mutant, while the gene for autolysin E (atlE) was not found among the DEGs (21–23). Expression of the pst operon and phoR, involved in inorganic phosphate metabolism, was upregulated in the ΔphoU2 mutant, while that of the phn ABC transporter, which participates in phosphonate metabolism, was downregulated. The expression of ATP synthase was upregulated (Table 2).
TABLE 2.
Genes differentially expressed between the ΔphoU2 mutant and the parent strain
| Gene function and gene | GenBank accession no. (location) | Description of product | Fold change in expression by: |
|
|---|---|---|---|---|
| RNA-Seq | RT-qPCRa | |||
| Growth | ||||
| yycF | NC_002976.3 (2591084–2591786) | DNA-binding response regulator YycF | 0.35 | 0.25 ± 0.08 |
| yycG | NC_002976.3 (2587909–2591072) | Sensor | 0.33 | 0.31 ± 0.14 |
| pflA | NC_002976.3 (2411377–2412133) | Pyruvate formate-lyase-activating enzyme | 0.14 | 0.12 ± 0.02 |
| nrdG | NC_002976.3 (2215127–2217511) | Anaerobic ribonucleoside triphosphate reductase-activating protein | 0.31 | ND |
| nrdD | NC_002976.3 (2215127–2217511) | Anaerobic ribonucleoside triphosphate reductase | 0.27 | 0.30 ± 0.14 |
| Biofilm formation | ||||
| icaR | NC_002976.3 (2333497–2334055) | Intercellular adhesion regulator | 4.58 | 4.20 ± 1.14 |
| serp0719 | NC_002976.3 (713668–716143) | Cell wall surface anchor family protein | 0.18 | 0.13 ± 0.04 |
| rsbU | NC_002976.3 (1724456–1725458) | Sigma factor B regulatory protein | 0.31 | ND |
| Autolysis | ||||
| serp1880 | NC_002976.3 (1904430–1905204) | Secretory antigen precursor SsaA | 3.56 | ND |
| serp1884 | NC_002976.3 (1909382–1909856) | Secretory antigen precursor SsaA | 9.47 | ND |
| serp2120 | NC_002976.3 (2144621–2145053) | Secretory antigen precursor SsaA-related protein | 1.92 | ND |
| serp2136 | NC_002976.3 (2161088–2161862) | Secretory antigen precursor SsaA | 3.03 | ND |
| serp0318 | NC_002976.3 (322333–323134) | LysM domain-containing protein | 1.86 | ND |
| Phosphate transport system | ||||
| serp0956 | NC_002976.3 (972985–973633) | Phosphate transport system regulatory protein PhoU | 5.10 | 48.95 ± 6.43 |
| serp0957 | NC_002976.3 (973639–974515) | Phosphate transporter ATP-binding protein | 3.75 | 67.96 ± 12.14 |
| serp0958 | NC_002976.3 (974602–975508) | Phosphate ABC transporter permease | 2.29 | ND |
| serp0959 | NC_002976.3 (975509–976436) | Phosphate ABC transporter permease | 3.32 | ND |
| serp0960 | NC_002976.3 (976642–977620) | Phosphate ABC transporter phosphate-binding protein | 16.46 | ND |
| serp2283 | NC_002976.3 (2323913–2325526) | Phosphonate ABC transporter permease | 0.31 | ND |
| serp2284 | NC_002976.3 (2323913–2325526) | Phosphonate ABC transporter permease | 0.31 | ND |
| serp2285 | NC_002976.3 (2325527–2326301) | Phosphonate ABC transporter ATP-binding protein | 0.26 | ND |
| serp2286 | NC_002976.3 (2326414–2327371) | Phosphonate ABC transporter substrate-binding protein | 0.27 | ND |
| serp0317 | NC_002976.3 (321130–322141) | Phosphate transporter family protein | 0.20 | 0.35 ± 0.09 |
| malA | NC_002976.3 (1114440–1116096) | Alpha-glucosidase | 0.38 | ND |
| lacD | NC_002976.3 (1838482–1839460) | Tagatose-1,6-diphosphate aldolase | 0.09 | ND |
| lacA | NC_002976.3 (1840938–1841367) | Galactose-6-phosphate isomerase subunit LacA | 0.06 | 0.13 ± 0.03 |
| lacG | NC_002976.3 (1834966–1836379) | 6-Phospho-beta-galactosidase | 0.24 | 0.33 ± 0.07 |
| galU | NC_002976.3 (2080817–2081684) | UTP-glucose-1-phosphate uridylyltransferase | 0.47 | ND |
| lacF | NC_002976.3 (1838148–1838463) | PTS system, lactose-specific IIA component | 0.12 | ND |
| serp2055 | NC_002976.3 (2078963–2080604) | Phosphoglucomutase/phosphomannomutase | 0.51 | ND |
| lacC | NC_002976.3 (1839463–1840396) | Tagatose-6-phosphate kinase | 0.09 | ND |
| Glycolysis/gluconeogenesis | ||||
| gntK | NC_002976.3 (2083340–2084882) | Gluconokinase | 0.19 | 0.33 ± 0.12 |
| pgi | NC_002976.3 (535899–537231) | Glucose-6-phosphate isomerase | 0.51 | ND |
| fruK | NC_002976.3 (355255–356931) | 1-Phosphofructokinase | 21.66 | ND |
| fbaA | NC_002976.3 (1771076–1771937) | Fructose-bisphosphate aldolase | 1.74 | ND |
| gapA2 | NC_002976.3 (1287162–1288188) | Glyceraldehyde-3-phosphate dehydrogenase | 0.62 | ND |
| pgk | NC_002976.3 (447676–448867) | Phosphoglycerate kinase | 0.35 | ND |
| gpmA | NC_002976.3 (2023859–2024546) | Phosphoglyceromutase | 0.23 | ND |
| ppdK | NC_002976.3 (2199426–2202054) | Pyruvate phosphate dikinase | 0.10 | 0.03 ± 0.01 |
| serp2133 | NC_002976.3 (2158590–2159589) | d-Lactate dehydrogenase | 1.54 | ND |
| serp2112 | NC_002976.3 (2135451–2136504) | Alcohol dehydrogenase | 0.03 | 0.38 ± 0.09 |
| serp2076 | NC_002976.3 (2100972–2102937) | Fructose-1,6-bisphosphatase | 0.39 | ND |
| Pentose phosphate pathway | ||||
| pgi | NC_002976.3 (535899–537231) | Glucose-6-phosphate isomerase | 0.51 | ND |
| xylB | NC_002976.3 (2123183–2124674) | d-Xylulose kinase | 0.63 | ND |
| gdh | NC_002976.3 (1867994–1868786) | Glucose-1-dehydrogenase | 3.52 | ND |
| deoC | NC_002976.3 (1784622–1785397) | Deoxyribose-phosphate aldolase | 0.62 | ND |
| prsA | NC_002976.3 (128258–129224) | Ribose-phosphate pyrophosphokinase | 0.62 | ND |
| tkt | NC_002976.3 (920891–922880) | Transketolase | 0.59 | ND |
| serp2076 | NC_002976.3 (2100972–2102937) | Fructose-1,6-bisphosphatase | 0.39 | ND |
| gntK | NC_002976.3 (2083340–2084882) | Gluconokinase | 0.19 | ND |
| deoB | NC_002976.3 (1781738–1783276) | Phosphopentomutase | 0.62 | ND |
| zwf-2 | NC_002976.3 (1111611–1113096) | Glucose-6-phosphate 1-dehydrogenase | 0.56 | ND |
| serp2055 | NC_002976.3 (2078963–2080604) | Phosphoglucomutase/phosphomannomutase | 0.51 | ND |
| rpe | NC_002976.3 (788507–789152) | Ribulose-phosphate 3-epimerase | 0.65 | ND |
| acnA | NC_002976.3 (932350–935056) | Aconitate hydratase | 0.64 | ND |
| icd | NC_002976.3 (1296194–1297463) | Isocitrate dehydrogenase | 0.46 | ND |
| serp2324 | NC_002976.3 (2360268–2361546) | Branched-chain alpha-keto acid dehydrogenase subunit E2 | 0.24 | ND |
| sucC | NC_002976.3 (815833–817000) | Succinyl coenzyme A synthetase subunit beta | 1.78 | ND |
| sdhB | NC_002976.3 (730805–733414) | Succinate dehydrogenase iron-sulfur subunit | 0.20 | ND |
| sdhA | NC_002976.3 (2118832–2119732) | Succinate dehydrogenase flavoprotein subunit | 0.17 | ND |
| fumC | NC_002976.3 (1444325–1445711) | Fumarate hydratase | 0.51 | ND |
| sucA | NC_002976.3 (1002484–1005289) | 2-Oxoglutarate dehydrogenase E1 | 0.44 | ND |
| sucB | NC_002976.3 (1001203–1002466) | Dihydrolipoamide succinyltransferase | 0.25 | ND |
| mqo-3 | NC_002976.3 (2350000–2351497) | Malate:quinone oxidoreductase | 0.43 | ND |
| mqo-1 | NC_002976.3 (1970555–1972034) | Malate:quinone oxidoreductase | 0.48 | ND |
| pckA | NC_002976.3 (1409888–1411481) | Phosphoenolpyruvate carboxykinase | 0.53 | ND |
| serp0857 | NC_002976.3 (868756–869623) | 2-Oxoglutarate ferredoxin oxidoreductase subunit beta | 0.56 | ND |
| serp0856 | NC_002976.3 (866995–868756) | Pyruvate ferredoxin oxidoreductase, alpha subunit | 0.44 | ND |
| serp2325 | NC_002976.3 (2361559–2362600) | Acetoin dehydrogenase, E1 component, beta subunit | 0.33 | ND |
| serp2327 | NC_002976.3 (2363665–2365018) | Acetoin dehydrogenase, E3 component, dihydrolipoamide dehydrogenase | 1.55 | ND |
| serp1076 | NC_002976.3 (1120885–1122205) | 2-Oxoisovalerate dehydrogenase E2 | 0.51 | ND |
| serp1077 | NC_002976.3 (1122217–1124192) | 2-Oxoisovalerate dehydrogenase E1 | 0.40 | ND |
| serp1078 | NC_002976.3 (1122217–1124192) | 2-Oxoisovalerate dehydrogenase E1 | 0.32 | ND |
| lpdA | NC_002976.3 (1124206–1125628) | 2-Oxoisovalerate dehydrogenase E3 | 0.34 | ND |
| serp2381 | NC_002976.3 (2428108–2431126) | NADH:flavin oxidoreductase/fumarate reductase, flavoprotein subunit | 0.02 | 0.082 |
qRT-PCR data are given as the means ± standard deviations of the results from three independent experiment. ND, not done.
Using KEGG analysis, the genes differentially expressed between the ΔphoU2 mutant and the parent strain were involved not only in phosphate metabolism, bacterial growth, and biofilm formation but also in various pathways or processes, such as the pentose phosphate pathway, galactose metabolism, the TCA cycle, glycolysis and gluconeogenesis, respiratory chain reactions, ABC transporter activity, the phosphotransferase (PTS) system, the urea cycle, and ribosome protein production (Table 2).
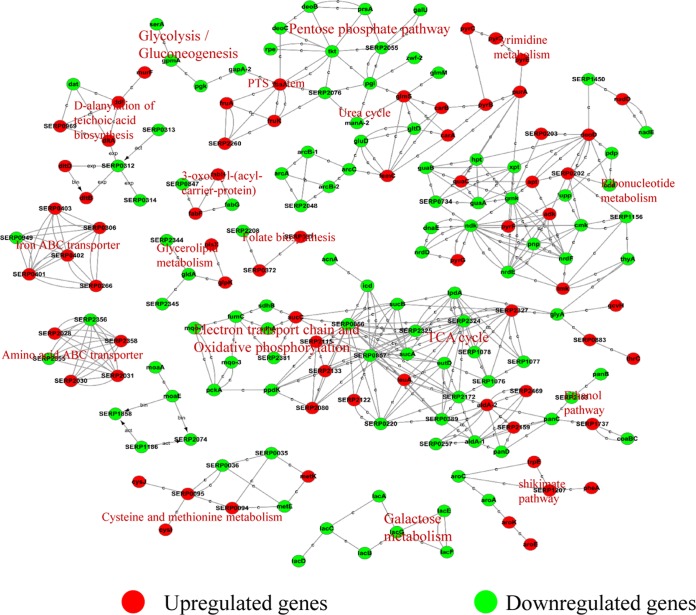
To further determine the links between the DEGs and the biological behavior in the ΔphoU2 mutant, a protein-protein interaction (PPI) network of DEGs based on the KEGG database (http://www.genome.jp/kegg/pathway.html) was constructed using Cytoscape software. The proteins encoded by DEGs were pooled with 174 proteins involved in various metabolic pathways. The expression of genes for proteins involved in the pentose phosphate pathway, galactose metabolism, and amino acid synthesis and TCA cycle genes was downregulated (Fig. 9). In the pentose phosphate pathway, NADP (NADPH) and pentose are important products. NADPH plays a crucial role in coping with oxidative stress (24–29). As pentose is utilized to synthesize DNA and RNA, the absence of DNA and RNA in the ΔphoU2 mutant leads to bacterial growth retardation.
FIG 9.
Interaction networks between DEGs identified by transcriptome comparison. The proteins encoded by DEGs (red, green), which were identified by comparison (at 6 h) of the transcriptomes of the ΔphoU2 mutant and the parent strain, were extracted to construct a protein-protein interaction network. The lines in the network represent protein-protein interactions, including binding/association, phosphorylation, activation, and inhibition. Proteins encoded by upregulated and downregulated DEGs are indicated in red and green, respectively.
Biological validation of genes differentially expressed between the ΔphoU2 mutant and the parent strain.
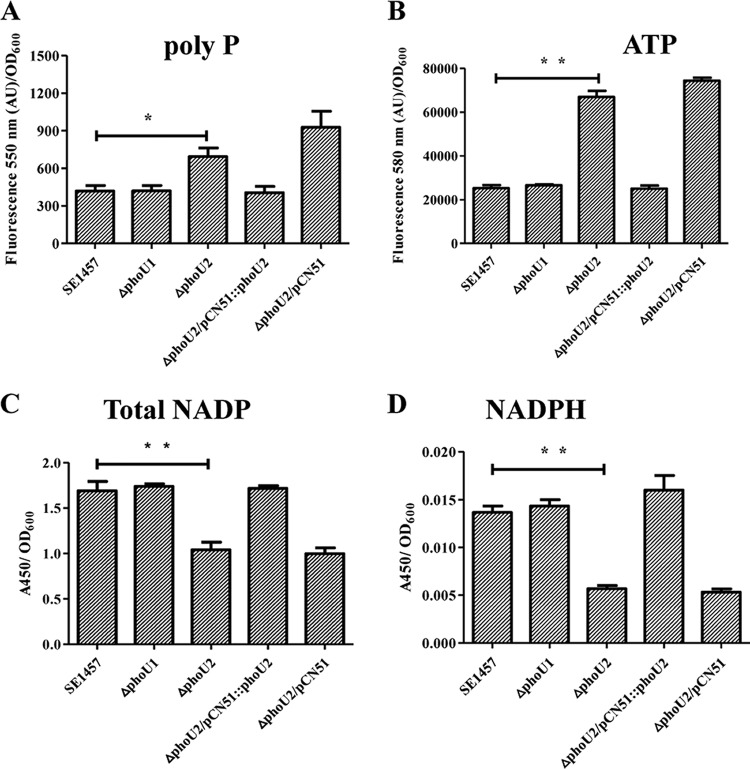
DEGs related to inorganic phosphate metabolism, ATP synthesis, and the pentose phosphate pathway were validated by the detection of intracellular polyP, ATP, and NADPH, respectively.
The expression of DEGs involved in inorganic phosphate metabolism, the pst operon, and serp0317 was upregulated in the ΔphoU2 mutant. Inorganic phosphate (Pi) taken up from the environment is used in bacterial metabolism, and the redundant inorganic phosphate is stored as a high-molecular-weight inorganic polyphosphate (polyP) in bacterial cells. To investigate the effects of the ΔphoU1 and ΔphoU2 deletions on Pi removal, we evaluated the levels of intracellular polyP in bacterial cells using a DAPI (4′,6-diamidino-2-phenylindole)-based fluorescence approach. The results showed that ΔphoU2 mutant cells accumulated higher levels of polyP (1.66-fold) than the parent strain (P < 0.05), while the amount of intracellular polyP in the ΔphoU1 mutant was similar to that in the parent strain (Fig. 10A). The polyP content in the phoU2-complemented ΔphoU2/pCN51::phoU2 strain was restored to that in the parent strain, whereas the transformation of plasmid pCN51 had no effect on polyP accumulation in the ΔphoU2 mutant.
FIG 10.
Intracellular polyP, ATP, NADP, and NADPH in the ΔphoU2 mutant. Bacteria were grown to exponential phase in TSB medium. PolyP was assessed by measuring the fluorescence emission of the DAPI-polyP complex at 550 nm. (A) The fluorescence (in arbitrary units [AU]) of the DAPI-polyP complex was measured at 550 nm to evaluate the amount of intracellular polyP. (B) The amount of ATP was determined by measuring the fluorescence emission of the ATP complex at 587 nm. Bacteria were grown to exponential phase in TSB medium (OD600 = 0.5). (C and D) The amounts of total NADP and NADPH were measured using an NADP/NADPH quantification kit (Sigma). The amounts of total NADP and NADPH were determined by measuring the absorbance of the NADPH complex at 450 nm. Data (means ± SDs) are from three independent experiments. **, P < 0.01; *, P < 0.05.
Since polyP in cells can be converted to ATP by phosphotransferases (30) and the transcription of ATP synthase was upregulated in the ΔphoU2 mutant, we determined the intracellular ATP levels in the ΔphoU2 mutant, the ΔphoU1 mutant, and the parent strain using an ATP colorimetric/fluorometric assay kit (Sigma). The intracellular ATP level in the ΔphoU2 mutant was 2.7-fold higher than that in the parent strain (P < 0.01) and the ΔphoU1 mutant (Fig. 10B). The amount of ATP recovered from the complemented ΔphoU2/pCN51::phoU2 strain was similar to that recovered from the parent strain.
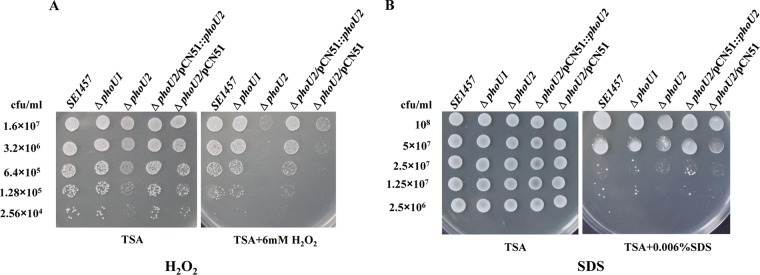
The expression of genes involved in the pentose phosphate pathway was downregulated in the ΔphoU2 mutant. Because most of the NADP was generated via PPP and intracellular NADPH, as a reduced form of NADP, plays a very important role in protecting against the toxicity of reactive oxygen species (ROS), intracellular NADPH levels in the ΔphoU2 mutant, the ΔphoU1 mutant, and the parent strain were determined using an NADP/NADPH quantification kit (Sigma). The total amount of NADP in the ΔphoU2 mutant (A450, 1.041 ± 0.085) was significantly reduced compared with that in the parent strain (A450, 1.691 ± 0.103) (P < 0.05) (Fig. 10C), while the amount of NADPH in the ΔphoU2 mutant (A450, 0.014 ± 0.001) was 3-fold lower than that in the parent strain (A450, 0.006 ± 0.001) (P < 0.01) (Fig. 10D). The amount of total NADP or NADPH in the phoU2-complemented ΔphoU2/pCN51::phoU2 strain was restored to the level in the parent strain. The reduced content of NADPH in the ΔphoU2 mutant compared with that in the parent strain may have led to the weakened capacity of the ΔphoU2 mutant to withstand oxidative stress. Then, the effects of phoU2 on the bacterial response to H2O2 were investigated. SDS was also determined to be another stress control. Different concentrations of the bacterial culture were spotted onto TSA containing 6 mM H2O2 or 0.006% SDS (8, 31). The ΔphoU2 mutant displayed higher sensitivity to H2O2, and no bacterial survival was detected when 6.4 × 105 CFU was spotted onto plates containing 6 mM H2O2 (Fig. 11A); bacteria of the parent strain were observed at 2.56 × 104 CFU. Complementation of the ΔphoU2 mutant could restore sensitivity to H2O2, the level of which reached that of the parent strain. However, the sensitivity of the ΔphoU1 mutant to H2O2 was similar to that of the parent strain. Both the ΔphoU1 mutant and the ΔphoU2 mutant displayed a level of sensitivity to SDS similar to that of the parent strain (Fig. 11B).
FIG 11.
Sensitivity of the ΔphoU1 and ΔphoU2 mutants to H2O2 and SDS. Overnight cultures of the bacterial strains were diluted 1:200 in fresh TSB medium and incubated at 37°C with aeration for 3 h until the OD600 was approximately 2. After 2-fold serial dilution, each 5-μl aliquot was spotted onto a TSB agar plate containing 6 mM H2O2 or 0.006% SDS and incubated overnight at 37°C. The growth of colonies on plates containing H2O2 or SDS was photographed.
DISCUSSION
PhoU in E. coli has been identified to be a regulator of phosphate uptake (32–34). Increasing attention has been focused on the regulation of PhoU in bacterial drug tolerance and the stress response. On the basis of the conserved motif of the PhoU protein, two PhoU gene homologs (serp0956 and serp0316) have been identified in S. epidermidis and named phoU1 and phoU2, respectively. phoU1 is located in the pst operon and shares a high degree of homology with phoU of E. coli. phoU2 (serp0316) is cotranscribed with serp0317 (see Fig. S1 in the supplemental material). serp0317 is another inorganic phosphate transport gene with homology to pit in E. coli (35, 36). Using bioinformatics analysis, the genome of S. aureus (NCTC8325) was found to contain two PhoU gene homologs (SAOUHSC_01384 and SAOUHSC_00669). phoU2 of S. epidermidis shares 86% identity with SAOUHSC_00669 at the nucleotide sequence level and PhoU2 shares 97% identity at the amino acid sequence level with SAOUHSC_00669 in S. aureus. SAOUHSC_00669 has been investigated as a gene required for the sensitivity of an S. aureus strain with a point mutation in pitA (SAOUHSC_00670) to daptomycin (11, 12).
The number of PhoU homologs varies in different bacterial species. In E. coli, P. aeruginosa, and Streptococcus pneumoniae, there is only one PhoU, while in T. maritima, M. tuberculosis, M. marinum, S. aureus, and S. epidermidis, two PhoU homologs have been identified. In E. coli, phoU plays an important role in polyP accumulation (34) and the formation of multidrug-resistant bacteria (5, 34). In P. aeruginosa, PhoU is a negative regulator of intracellular ppGpp and polyP. However, phoU mutation has no effect on P. aeruginosa biofilm formation (8). In species of mycobacteria, there are two phoU homologs, phoY1 and phoY2. phoY2 has been investigated as the functional homolog of phoU, regulating the generation of multidrug-tolerant bacteria and maintaining metabolic homeostasis and adaptation to stress conditions in M. tuberculosis and M. marinum (6, 7).
Alignment of the amino acid sequences of PhoU1 and PhoU2 from S. epidermidis RP62A, PhoU from E. coli K-12, and the two PhoU homologs (PhoY1 and PhoY2) from M. tuberculosis H37Rv showed that the metal ion-binding sites are conserved in all the PhoU homologs (Fig. S2). To investigate the regulatory functions of the phoU homologs, the ΔphoU1 and ΔphoU2 mutants of the S. epidermidis 1457 strain were constructed and the transcriptomes of the deletion mutants and the parent strain were compared. We detected the transcription levels of phoU1 (SERP0956) and phoU2 (SERP0316) in SE1457 by RT-qPCR at different time point (4 h, 6 h, 8 h, 10 h, and 12 h), and both phoU1 and phoU2 of SE1457 were expressed at the time points evaluated (Fig. S1). In logarithmic phase (6 h), 945 genes were differentially expressed between the ΔphoU2 mutant and the parent strain; among these, 474 were upregulated and 471 were downregulated. However, only 92 DEGs were detected between ΔphoU1 and the parent strain during logarithmic phase and 2 DEGs were detected during stationary phase. On the basis of Gene Ontology analysis of the genes differentially expressed between the ΔphoU1 mutant and the parent strain, 23 DEGs that were downregulated in the ΔphoU1 mutant were involved in translation, ribosomal structure, and biogenesis, while 69 upregulated DEGs were involved in 17 pathways or metabolic processes. However, the differences in the levels of expression of none of these DEGs were statistically significant (P > 0.05). It is worth mentioning that the pst operon was not identified among the genes differentially expressed between the ΔphoU1 mutant and the parent strain, and the results were validated by RT-qPCR. Hence, in this study, we focused on investigating phoU2 functions in S. epidermidis.
The ΔphoU2 mutant of S. epidermidis displayed growth retardation under both microaerobic and oxic conditions, in accordance with the findings in an E. coli strain with a PhoU deletion (37). The deletion of phoU1 had no effect on bacterial growth. Analysis of the DEGs (genes differentially expressed between the ΔphoU2 mutant and the parent strain) showed that in the ΔphoU2 mutant, the expression of pflA and nrdDG was downregulated. These genes are involved in the growth of S. epidermidis under microaerobic conditions. PflA is an enzyme activating PflB, a pyruvate formate-lyase that catalyzes the reversible conversion of pyruvate to formate, thereby producing acetyl coenzyme A. Thus, PflA plays an important role in utilization of the energy supply when pyruvate is available and favors the growth of cells under fermentation conditions (18). The protein encoded by nrdDG is a class III ribonucleotide reductase that catalyzes the synthesis of deoxynucleoside triphosphates (dNTPs) via the reduction of nucleoside triphosphates under anaerobic conditions (38–40). In the ΔphoU2 mutant, the expression of yycFG is significantly downregulated (P < 0.01); yycFG is an essential two-component system in Gram-positive bacteria that regulates cell wall metabolism, cell division, virulence, and biofilm formation (41, 42). Expression of the essential two-component system yycFG was downregulated during the logarithmic phase but was not detected during the stationary phase. This finding is in accordance with the results of the assessments of the ΔphoU2 mutant under the different growth conditions. In addition, the downregulated expression of DEGs involved in the TCA cycle, glycolysis and gluconeogenesis, and the pentose phosphate pathway may promote growth retardation (Fig. 12 and Table 3). The growth retardation of the ΔphoU2 mutant did not persist to stationary phase, as there were no differences in the CFU counts from those of the parent strain in stationary phase.
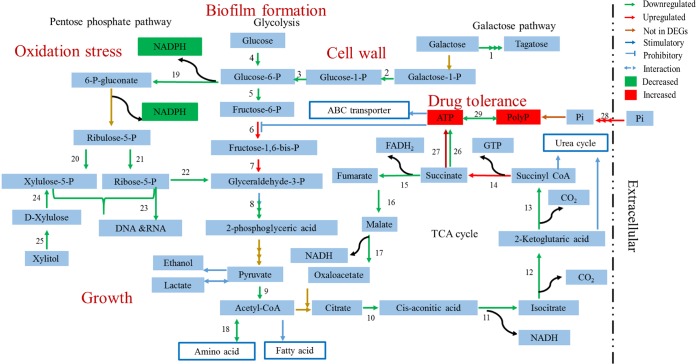
FIG 12.
Major metabolic pathways in the ΔphoU2 mutant revealed by DEGs. Shown are transcription of enzyme-encoding genes that were downregulated and upregulated, enzyme-encoding genes that were not found in DEGs (genes differentially expressed between the ΔphoU2 mutant and the parent strain), stimulatory reactions, prohibitory reactions, reversible reaction interactions between the two products, and products whose amounts were increased or decreased. The numbers represent the enzyme-encoding genes listed in Table 3. P, phosphate; CoA, coenzyme A.
TABLE 3.
DEGs involved in major metabolic pathway
| DEG no. | Gene name(s) | Enzyme(s) encoded |
|---|---|---|
| 1 | lacA, lacD, lacG | Galactose-6-phosphate isomerase subunit LacA, tagatose-1,6-diphosphate aldolase, 6-phospho-beta-galactosidase |
| 2 | galU | UTP-glucose-1-phosphate uridylyltransferase |
| 3 | serp2055 | Phosphoglucomutase/phosphomannomutase |
| 4 | gntK | Gluconokinase |
| 5 | pgi | Glucose-6-phosphate isomerase |
| 6 | fruK | 1-Phosphofructokinase |
| 7 | fbaA | Fructose-bisphosphate aldolase |
| 8 | gapA2, pgk, gpmA | Glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, phosphoglyceromutase |
| 9 | serp0856, serp0857 | 2-Oxoglutarate ferredoxin oxidoreductase subunit beta, pyruvate ferredoxin oxidoreductase, alpha subunit |
| 10 | acnA | Aconitate hydratase |
| 11 | acnA | Aconitate hydratase |
| 12 | icd | Isocitrate dehydrogenase |
| 13 | serp2324 | Branched-chain alpha-keto acid dehydrogenase subunit E2 |
| 14 | sucC | Succinyl coenzyme A synthetase subunit beta |
| 15 | sdhA, sdhB | Succinate dehydrogenase iron-sulfur subunit, succinate dehydrogenase flavoprotein subunit |
| 16 | fumC | Fumarate hydratase |
| 17 | mqo-1, mqo-3 | Malate:quinone oxidoreductase, malate:quinone oxidoreductase |
| 18 | serp1076, serp1077, serp1078, lpdA | 2-Oxoisovalerate dehydrogenase E2, 2-oxoisovalerate dehydrogenase E1, 2-oxoisovalerate dehydrogenase E1, 2-oxoisovalerate dehydrogenase E3 |
| 19 | zwf-2 | Glucose-6-phosphate 1-dehydrogenase |
| 20 | rpe | Ribulose-phosphate 3-epimerase |
| 21 | deoB | Phosphopentomutase |
| 22 | deoC | Deoxyribose-phosphate aldolase |
| 23 | prsA | Ribose-phosphate pyrophosphokinase |
| 24 | xylB | d-Xylulose kinase |
| 25 | tkt | Transketolase |
S. epidermidis is an important nosocomial pathogen that forms biofilms on implanted medical devices. We studied the regulatory role of phoU2 in this respect. The ΔphoU2 mutant exhibited impaired biofilm formation under both static and hydrodynamic conditions, in contrast to the findings obtained for P. aeruginosa. A study performed in 2015 found that inactivation of phoU had no effect on the formation of bacterial biofilms in P. aeruginosa (8). In the ΔphoU2 mutant, the expression of the sigma factor B regulator gene (rsbU), involved in biofilm formation, is downregulated (P < 0.01) (20). In addition, the downregulation of genes that participate in galactose metabolism plays an essential role in Bacillus subtilis biofilm formation. The intermediate product of galactose metabolism, UDP-galactose, is used in the synthesis of exopolysaccharide (EPS) of the biofilm matrix in B. subtilis (43). In accordance with these phenomena, the production of Aap was decreased in the ΔphoU2 mutant. Defective attachment of the ΔphoU2 mutant could be another important reason for the reduced biofilm formation. The decreased biofilm formation was not due to growth retardation, as the number of CFU of the ΔphoU2 mutant during the stationary phase was similar to that of the parent strain, and the time required for biofilm formation was extended to 48 h.
The downregulated expression of genes involved in the TCA cycle, glycolysis, and gluconeogenesis resulted in a lack of polysaccharide, which was also responsible for the biofilm formation defect in the ΔphoU2 mutant.
The ΔphoU2 mutant of S. epidermidis displayed a rough cell wall and rapid autolysis. The rough cell wall may indicate that the cell wall of the ΔphoU2 mutant was incomplete, potentially due to the low expression levels of the galactose metabolism genes. The intermediate products of galactose metabolism are involved in the synthesis of peptidoglycan. Peptidoglycan, which is also known as murein, is the most important component of Gram-positive bacterial cell walls (Fig. 12). In addition to the autolysis genes, ssaA and serp0318 were upregulated in the ΔphoU2 mutant; the incomplete cell wall may have led to the rapid autolysis in the ΔphoU2 mutant (22, 23). Besides the peptidoglycan, the level of another component of the cell wall, teichoic acid, may also have been decreased in the ΔphoU2 mutant. Teichoic acids are special components in the cell wall of most Gram-positive bacteria, such as species in the genera Staphylococcus, Streptococcus, and Bacillus. Teichoic acids are bacterial copolymers of glycerol phosphate or ribitol phosphate and carbohydrates linked via phosphodiester bonds and are the essential product of glycolysis or the pentose phosphate pathway. The downregulation of glycolysis or the pentose phosphate pathway in the ΔphoU2 mutant may reduce the level of production of teichoic acids, which would result in a rough cell surface and an increase in autolysis capacity (44, 45).
Inorganic phosphate (Pi) is an important macronutrient for all living organisms, comprising up to 3% of the bacterial dry weight, and is involved in many important pathways (8). The expression of genes in the pst operon and serp0317 was upregulated in the ΔphoU2 mutant, which is consistent with the intracellular accumulation of polyP in the ΔphoU2 mutant. A portion of the inorganic phosphate taken up from the environment was used for bacterial metabolism, and redundant inorganic phosphate was stored as high-molecular-weight inorganic polyphosphate (polyP) in bacterial cells (46) (Fig. 12). PolyP is a linear polymer composed of several molecules of orthophosphate (Pi) that are linked by energy-rich phosphoanhydride bonds (30). PolyP is a known stress response molecule that accumulates in microorganisms in response to nutrient deprivation, high salt concentrations, or other environmental stress conditions (30, 47). An imbalance in the amount of intracellular polyP in E. coli elicits a defective response to oxidative, osmotic, and thermal stresses (5, 34). In M. tuberculosis, a phoY2 transposon insertion mutant accumulates large amounts of polyP and has an increased sensitivity to thermal stress, H2O2, and antibiotics (6, 7). The characteristics of phosphate metabolism in the S. epidermidis ΔphoU2 mutant are similar to those in phoU deletion mutants or insertion inactivation mutants of E. coli, P. aeruginosa, M. tuberculosis, and M. marinum (5–8).
To study the regulation of phosphate metabolism by PhoU1 and PhoU2, we determined the intracellular inorganic phosphate content and the growth of the PhoU mutants (the ΔphoU1 and ΔphoU2 mutants), in addition to the amount of polyP, under phosphate-limiting conditions. There were no differences in the intracellular inorganic phosphate content between the PhoU mutants and the parent strain (SE1457) (Fig. S3B). The growth curves of the PhoU mutants under Pi-limiting conditions were similar to those of the mutants cultured in TSB medium (Pi-replete conditions) (Fig. S3A). Comparison of the transcriptomes of the PhoU mutants and the parent strain showed that the transcription of the pst operon was upregulated in the ΔphoU2 mutant, which suggests that phosphate uptake was increased in the ΔphoU2 mutant and the extra phosphate was utilized to synthesize polyP to maintain the intracellular inorganic phosphate at a level similar to that in SE1457. However, in the ΔphoU1 mutant there was no difference in the level of pst operon expression, the intracellular inorganic phosphate and polyP content, or growth under Pi-limiting conditions compared with the findings for SE1457. These results suggest that PhoU2 but not PhoU1 plays an important role in the homeostasis of intracellular Pi.
The polyP in bacterial cells could be converted to ATP by phosphotransferases. In addition, among the genes differentially expressed between the ΔphoU2 mutant and the parent strain, the expression of genes encoding ATP synthase was upregulated. Thus, we verified that the levels of intracellular ATP were higher in the ΔphoU2 mutant strain than the parent strain, which is consistent with the results obtained for M. marinum (7). ATP is the most important energy-providing substrate. A high level of intracellular ATP influences the expression of many genes and metabolic pathways, such as the genes for ATP-binding cassette (ABC) transporters. ABC transporters utilize the energy from ATP binding and hydrolysis to transport various substrates, including ions, amino acids, peptides, sugars, and other molecules, across cellular membranes (48). Excessive ABC transporter activity would result in an imbalance in the uptake of substances. A high level of ATP would maintain cellular metabolism at an unhealthy active level (49). Under this condition, bacteria are easily killed by bactericidal antibiotics. Therefore, S. epidermidis ΔphoU2 showed a reduced drug tolerance compared with the wild-type strain (Fig. 12).
On the basis of the protein-protein interaction network developed for the proteins encoded by the DEGs, 174 proteins encoded by DEGs were found to be involved in various metabolic pathways, such as the pentose phosphate pathway, the TCA cycle, and galactose metabolism. Among these, the expression of all genes that participate in the pentose phosphate pathway was downregulated in the ΔphoU2 mutant. We found that the amount of NADPH in the ΔphoU2 mutant decreased 3-fold compared with that in the parent strain. NADPH is produced mainly via the pentose phosphate pathway, which parallels the glycolysis pathway. The pentose phosphate pathway generates NADPH and pentoses (5-carbon sugars) as well as ribose-5-phosphate. NADPH provides the reducing equivalents for biosynthetic reactions and the oxidation-reduction involved in protection against the toxicity of reactive oxygen species (ROS), allowing the regeneration of glutathione (GSH) (24–29). The decreased NADPH in the ΔphoU2 mutant corresponded to its high sensitivity to H2O2 (Fig. 12). The major product of PPP is ribose-5-phosphate, a precursor for the synthesis of nucleotides (DNA and RNA). The reduced synthesis of ribose-5-phosphate led to a lack of DNA and RNA, and this reduced amount of ribose-5-phosphate might be responsible for the growth retardation seen in the ΔphoU2 mutant (Fig. 12).
To confirm the role of phoU2 on the regulation of growth and biofilm formation in SE1457, we used the antisense technology to test the effect of silencing of phoU2 expression on the ΔphoU1 mutant. First, we constructed a plasmid which could express phoU2 antisense RNA (AS-phoU2) in the presence of 250 ng/ml anhydrotetracycline (ATc), and the plasmid was transferred to the ΔphoU1 mutant to obtain the AS-phoU2 ΔphoU1 mutant. After induction with ATc, the transcription level of phoU2 was reduced to 10% in the AS-phoU2 ΔphoU1 mutant compared with that in the ΔphoU1 mutant. The AS-phoU2 ΔphoU1 mutant displayed a level of growth retardation that was the same as that in the ΔphoU2 mutant (Fig. S4A), while the level of biofilm formation was less than that in the ΔphoU1 mutant carrying the vector pMX6 (P < 0.01) (Fig. S4B).
In summary, phoU2 regulates growth, biofilm formation, oxidative stress, and drug tolerance via some important pathways and processes, including the pentose phosphate pathway, glycolysis, the TCA cycle, inorganic phosphate metabolism, galactose metabolism, and ABC transporter activity in SE1457. However, no obvious phenotype of the ΔphoU1 mutant was observed in the presence of phoU2 in SE1457. The mechanism of phoU2 regulation and the interaction between phoU1 and phoU2 in SE1457 warrant further investigation.
The effects of PhoU1 and PhoU2 on biofilm formation and antibiotic tolerance in S. epidermidis clinical strains need to be further studied by the use of gene knockout or antisense RNA technology.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and chemicals.
All of the bacterial strains and plasmids used in the study are shown in Table 4. The biofilm-positive strain S. epidermidis ATCC 35984 (RP62A; GenBank accession number NC_002976) and non-biofilm-forming strain ATCC 12228 (GenBank accession number NC_004461) were purchased from the American Type Culture Collection (ATCC; Manassas, VA) (50). S. epidermidis 1457 (SE1457) and S. aureus RN4220 were provided by Gao Fu from the University of Hong Kong. The S. epidermidis strains and S. aureus RN4220 were cultured in tryptic soya broth (TSB; Oxoid, Basingstoke, UK) at 37°C with shaking at 220 rpm. Glucose was added to the TSB medium at a concentration of 0.5% for the detection of biofilm formation. Electroporation was used for plasmid transformation, and B2 medium (1% casein hydrolysate, 2.5% yeast extract, 0.5% glucose, 2.5% NaCl, 0.1% K2HPO4, pH 7.5) was used for the recovery of bacteria. The antibiotics used in this study were purchased from Sigma Chemical Co. (Los Angeles, CA, USA) and used at concentrations of 10 mg/liter for chloramphenicol, 100 mg/liter for ampicillin, 50 ng/ml for anhydrotetracycline, and 10 mg/liter for erythromycin.
TABLE 4.
Bacterial strains and plasmids used in the present study
| Bacterial strain or plasmid | Description | Source |
|---|---|---|
| Bacterial strains | ||
| S. epidermidis 1457 | Clinical strain, biofilm positive | |
| ΔphoU1 mutant | phoU1 deletion mutant obtained using SE1457 as the parent strain | This study |
| ΔphoU2 mutant | phoU2 deletion mutant obtained using SE1457 as the parent strain | This study |
| ΔphoU2/pCN51::phoU2 | phoU2 mutant complemented with plasmid pCN51 harboring the phoU2 gene | This study |
| ΔphoU2/pCN51 | phoU2 mutant complemented with plasmid pCN51 | This study |
| S. epidermidis RP62A | Standard strain, biofilm positive | ATCC |
| S. epidermidis ATCC 12228 | Standard strain, biofilm negative | ATCC |
| S. aureus RN4220 | Restriction-deficient strain permitting shuttle of a plasmid modified by its host specificity determination from Gram-negative to Gram-positive bacteria | Gao Fu, University of Hong Kong |
| Plasmids | ||
| pKOR1 | Temp-sensitive E. coli (Ampr)-Staphylococcus (Cmr) shuttle vector | Li Min, Institute of Antibiotics, Huashan Hospital |
| pKOR1-ΔphoU1 | Recombinant plasmid | This study |
| pKOR1-ΔphoU2 | Recombinant plasmid | This study |
| pCN51 | A Cd2+-inducible shuttle plasmid, Ermr | Wageningen University, Holland |
| pCN51- phoU2 | A Cd2+-inducible shuttle plasmid, Ermr; the phoU2 gene was cloned into plasmid pCN51 | This study |
Construction of ΔphoU1 and ΔphoU2 mutants and complemented strains.
The phoU1 and phoU2 deletion mutants of the SE1457 strain were constructed by allelic replacement using the temperature-sensitive plasmid pKOR1 as described by Bae and Schneewind (51). Briefly, the upstream and downstream fragments of phoU1 and phoU2 were amplified by PCR and separately cloned into the pKOR1 vector, resulting in pKOR1-ΔphoU1 and pKOR1-ΔphoU2. The recombinant plasmid pKOR1-ΔphoU was successively transferred into E. coli DH5α, S. aureus RN4220, and then SE1457, followed by allelic replacement as described previously (52, 53). The phoU1and phoU2 deletion mutants were verified by PCR, RT-qPCR, and direct sequencing and are referred to as the ΔphoU1 and ΔphoU2 mutants. The complemented ΔphoU2 mutant strain was constructed using a shuttle vector, pCN51 (54). The phoU2 gene and its Shine-Dalgarno region were amplified by PCR using primers phoU2-BamHI-F/phoU2-KpnI-R, whose sequences are provided in Table 5. Plasmid pCN51-phoU2 was used for complementation and was constructed by inserting a fragment of the digested PCR products of the phoU2 gene with BamHI and KpnI (55). Plasmid pCN51-phoU2 was transformed by electroporation into the ΔphoU2 mutant, forming the complemented ΔphoU2/pCN51::phoU2 strain. The ΔphoU2 strain containing the empty vector pCN51 was designated the ΔphoU2/pCN51 mutant. The primers used in this assay are listed in Table 5.
TABLE 5.
Primers used in this study
| Primer use and primera | Primer sequence (5′–3′) | Locationb | Size of PCR product (bp) | Notec |
|---|---|---|---|---|
| Construction of ΔphoU1 mutant | ||||
| phoU1 us-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTAAATGCCTCAAGCAGAATTC | 974705–974724 | 1,061 | attB1 |
| phoU1us-R | GGGGTACCAATTGCCATTGCATCTTATCC | 973625–973645 | KpnI | |
| phoU1ds-F | GGGGTACCGATTAAATTATCAAATCC TATTG | 972969–972991 | 823 | KpnI |
| phoU1ds-R | GGGGACCACTTTGTACAAGAAAGCTGGGTTTGCTCAGAATAAAGGAAAAG | 972127–972147 | attB2 | |
| Construction of ΔphoU2 mutant | ||||
| phoU2 us-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTGTTGATCGTGGTAGACCG | 319402–319419 | 1,100 | attB1 |
| phoU2 us-R | GGGGTACCCATTAAAAATCC TCCATTTTGA | 320480–320501 | KpnI | |
| phoU2 ds-F | GGGGTACCTAAGGGAGTCTTTATTTATGTC | 321114–321135 | 959 | KpnI |
| phoU2 ds-R | GGGGACCACTTTGTACAAGAAAGCTGGGTCACCCATGTTACAACCATAC | 322053–322072 | attB2 | |
| Construction of ΔphoU2 complemented strain | ||||
| phoU2-BamHI-F | CGCGGATCCGTAAATCAGTTCCTCA | BamHI | ||
| phoU2-KpnI-R | CGGGGTACCTAAATAAAGACTCCCT | KpnI | ||
| RT-qPCR | ||||
| gyrB F | AGAAGAGGAAGTTAGAGAAGA | 2611073–2611093 | 168 | |
| gyrB R | GCATATCC ACTGTTATATTGAAG | 2610926–2610948 | ||
| phoU1 F | CGTCTTGGTCTTCGTGTA | 973556–973573 | 169 | |
| phoU1 R | CAATAGGTTGTTGTCTCGTAAT | 973405–973426 | ||
| phoU2 F | GCTGTAGGATTACTTGTAGAC | 320874–320894 | 200 | |
| phoU2 R | GCTTGACACTTATCTGCTATT | 321073–321053 |
Primers were designed according to the genomic sequence of S. epidermidis RP62A (GenBank accession number NC_002976). F, forward primer; R, reverse primer.
Location of the primer in the genomic sequence of S. epidermidis RP62A.
The underlined sequences represent the BP reaction sites or restriction enzyme sites.
Growth curves of the ΔphoU1 and the ΔphoU2 mutant strains.
The SE1457, ΔphoU1, ΔphoU2, ΔphoU2/pCN51::phoU2, and ΔphoU2/pCN51 strains were cultured in TSB at 37°C with shaking for 12 h. Overnight cultures of the S. epidermidis strains were diluted (1:200) in 10 ml TSB in a 100-ml flask and incubated at 37°C under aerobic or microanaerobic conditions with shaking at 220 rpm. The OD600 was measured using a UV spectrophotometer (Eppendorf, Hamburg, Germany) (53).
Morphology of the ΔphoU1 and ΔphoU2 mutants observed by TEM.
The SE1457, ΔphoU1, ΔphoU2, ΔphoU2/pCN51::phoU2, and ΔphoU2/pCN51 strains were cultured in TSB at 37°C for 6 h. The log-phase bacteria were rinsed with phosphate-buffered saline (PBS), prefixed with 2.5% glutaraldehyde at 4°C for 2 h, and then fixed in 1% osmium for 3 h. The fixed samples were dehydrated in a graded ethanol series, embedded in epoxy resin, and stained with uranyl acetate and lead citrate. Ultrathin sections were cut with a Leica Ultracut microtome and observed under a transmission electron microscope (TEM; Philips Tecnai-12 Biotwin). TEM micrographs were separately obtained at magnifications of ×11,500 and ×26,500 (56, 57).
Microtiter plate assay of biofilm formation.
The biofilm-forming ability of the strains was assessed using a semiquantitative microtiter plate assay, based on the protocol described by Christensen et al. (58). Briefly, overnight cultures of the S. epidermidis strains were diluted (1:200) into fresh TSB (containing 0.5% glucose). Aliquots of the diluted cultures were inoculated into polystyrene 96-well flat-bottomed microtiter plates (Costar; Corning, USA) and incubated at 37°C for 6 h, 12 h, 24 h, or 48 h. After the planktonic cultures were decanted, the plates were gently rinsed three times with PBS. The biofilms were fixed with 99% methyl alcohol for 15 min, stained with 2% crystal violet for 5 min, rinsed with running tap water, and air dried at room temperature. The optical densities at 570 nm (OD570s) of the plates were then determined using a spectrophotometer (DTX880; Beckman Coulter, Fullerton, CA) (52). The biofilm formation by each strain was assessed three times using strains ATCC 12228 and ATCC 35984 as non-biofilm-forming and biofilm-forming controls, respectively.
CLSM of biofilms.
An overnight culture of each S. epidermidis strain (1:200 dilution) was inoculated into 2 ml TSB (with 0.5% glucose supplementation) in a cell culture dish containing a glass coverslip (World Precision Instruments, USA) (59, 60). After culturing under static conditions at 37°C for 24 h, the dish was gently washed three times with saline and then stained with LIVE/DEAD reagents (1 μM SYTO9 and 1 μM propidium iodide [PI]; Thermo Fisher Scientific, Houston, TX) for 20 min and observed using a confocal laser scanning microscope (CLSM; TCS-SP5, Leica, Germany) with a 63× oil immersion objective (numerical aperture, 1.4). Photomicrographs of the biofilms were generated using Leica LAS AF software. Three-dimensional images were created using IMARIS (version 7.0.0) software (Bitplane). The live and dead bacteria were quantified using ImageJ software. Three independent experiments were performed.
Flow-based biofilm assays.
A BioFlux 1000 system (Fluxion Biosciences) with a Leica microscope and temperature-controlled housing was used for all imaging experiments. Automated microscopy and image processing were performed using BioFlux Montage software. BioFlux 48-well plates (catalog no. 910-0047; Fluxion Biosciences) allowing up to 24 individual treatment conditions were primed with TSB from the inlet well at a shear setting of 2 dynes/cm2 for 10 min. Cultures of S. epidermidis were grown to mid-log phase and normalized to an OD600 of 0.15. Bacteria were seeded from the outlet well into the channel and viewing window at a shear setting of 2 dynes/cm2 for 3 s. After 1 h of incubation at 37°C, TSB was set to flow from the inlet well at a shear setting of 0.15 dyne/cm2 for the duration of the experiment. Images were automatically acquired every 10 min at multiple stage positions using bright-field illumination; images were also acquired in the red channel (tetramethyl rhodamine isothiocyanate [TRITC] filter set; catalog no. 86013v2; Chroma, Bellows Falls, VT) using a 200-ms exposure time. The background-corrected average pixel intensity per image was used to quantify biofilm formation (61).
Autolysis of the ΔphoU1 and ΔphoU2 mutants induced by Triton X-100.
Autolysis assays were performed as described by Brunskill and Bayles with minor modifications (62). Briefly, the S. epidermidis strains were cultured in TSB (containing 1 M NaCl) to mid-exponential phase (6 h) and harvested by centrifugation (4,000 × g, 4°C, 15 min). After the bacterial pellets were washed with ice-cold water, they were resuspended to an OD600 of 1.0 in lysis buffer (50 mM Tris-HCl buffer, pH 8.0, containing 0.05% Triton X-100) and incubated at 30°C with shaking at 200 rpm. The optical density at 600 nm was measured at 30-min intervals for 3 h (59).
Initial adherence capacity of the ΔphoU1 and ΔphoU2 mutants.
Primary attachment of the ΔphoU1 and ΔphoU2 strains to a polystyrene surface was assessed as described previously, with modifications (52, 53, 59). Briefly, overnight cultures of the SE1457, ΔphoU1, ΔphoU2, ΔphoU2/pCN51::phoU2, and ΔphoU2/pCN51 strains were inoculated into TSB and cultured at 37°C to logarithmic phase (OD600, ≈0.6). The bacterial culture was adjusted to an OD600 of 0.1 with PBS and inoculated into six-well plates (2 ml/well; Nunc, Roskilde, Denmark). After incubation at 37°C for 2 h, the plates were washed gently with PBS and observed under a microscope using a 40-fold objective lens. The numbers of attached cells in each photomicrograph were counted using ImageJ software, and three microscopic fields were observed per sample.
Assay of PIA in biofilms.
Polysaccharide intercellular adhesion (PIA) in the biofilms of the ΔphoU1 and ΔphoU2 mutants was semiquantified by a dot blot assay with a wheat germ agglutinin (WGA)-horseradish peroxidase (HRP) conjugate as described previously (63, 64). Briefly, overnight cultures of the S. epidermidis strains were inoculated into six-well plates (Nunc) and incubated at 37°C for 24 h. Biofilms were scraped off the bottoms of the wells, resuspended in 0.5 M EDTA, and centrifuged (13,000 × g, 5 min) after they were heated at 100°C for 5 min. The supernatant was treated with proteinase K (20 mg/ml) at 37°C for 3 h and inactivated at 100°C for 5 min. Serial dilutions of the PIA assay extract were transferred to a nitrocellulose membrane (Millipore, Billerica, MA) using a 96-well dot blot device (Biometra GmbH, Gottingen, Germany). The air-dried membrane was blocked with 5% (wt/vol) skim milk and subsequently incubated with the WGA (3.2 μg/ml)-HRP conjugate for 1 h (Lectinotest Laboratory, Lviv, Ukraine). HRP activity was visualized by chromogenic detection using 4-chloride-1-naphthol (Sigma) as the substrate. Quantitation of PIA was represented as the highest dilution of the supernatant in which HRP was detectable.
Detection of Aap.
Accumulation-associated protein (Aap) expression by the ΔphoU1 and ΔphoU2 strains was determined by Western blotting with an Aap-specific monoclonal antibody (MAb18B6) that was generated in our laboratory (60). Briefly, 24-h-old biofilms of the S. epidermidis strains were collected and adjusted to an identical OD600 after they were washed with PBS. The bacteria were treated with lysostaphin (Sigma) and centrifuged (20,000 × g) at 4°C for 30 min. The supernatants were separated using SDS-PAGE (7%) and blotted onto a polyvinylidene fluoride membrane (pore size, 0.45 μm; Millipore) by electrotransfer. The membrane was incubated with MAb18B6 (10 ng/ml) and then with goat anti-mouse IgG conjugated to HRP (Santa Cruz, Santa Cruz, CA), followed by visualization using an enhanced chemiluminescence (ECL) Western blotting system (Thermo Fisher Scientific, Waltham, MA).
Quantification of eDNA.
The isolation of extracellular DNA (eDNA) from the biofilms was performed as described previously (52, 53). Briefly, 24-h-old biofilms cultured in a 96-well polystyrene plate were chilled at 4°C for 1 h, and EDTA was added at a final concentration of 2.5 mM. After measurement of the OD600 of the unwashed biofilm (biofilm biomass), eDNA extraction solution (50 mM Tris-HCl, 10 mM ETDA, 500 mM NaCl, pH 8.0) was added to the wells. The biofilms were scraped off and centrifuged (13,000 × g) for 5 min at 4°C. The eDNA in the supernatant was extracted with phenol-chloroform-isoamyl alcohol (25:24:1), precipitated with 100% alcohol, and resuspended in Tris-EDTA buffer. The amount of eDNA was quantified by qPCR with SYBR Premix Ex Taq (TaKaRa Bio, Inc., Shiga, Japan) using the primers specific for gyrB (gyrase B gene), serp0306 (ferrichrome transport ATP-binding protein A gene), leuA (2-isopropylmalateynthase gene), and lysA (diaminopimelate decarboxylase A gene). Each gene was assayed in the qPCR in triplicate in three independent experiments. Relative quantitation of the eDNA in each sample was calculated as the total amount of eDNA (in nanograms) divided by the biofilm biomass (OD600).
Measurement of intracellular polyP in the ΔphoU1 and ΔphoU2 mutants.
A DAPI (4′,6-diamidino-2-phenylindole)-based fluorescence approach was used to evaluate the amount of intracellular polyP in the S. epidermidis cell suspensions as described previously (65). Logarithmic-phase cultures of the S. epidermidis strains grown in TSB were collected by centrifugation at 4,000 × g for 5 min at room temperature. The pellets were washed three times with 100 mM Tris-HCl (pH 7.4) and resuspended in the same buffer to an OD600 of 0.2. DAPI (Beyotime) was added to a final concentration of 10 μM. The DAPI fluorescence spectra (excitation, 415 nm; emission, 450 to 650 nm) were recorded using a Cary Eclipse fluorescence spectrophotometer (Varian) after 5 min of agitation at room temperature in the dark. The fluorescence (in arbitrary units) of the DAPI-polyP complex was measured at 550 nm for evaluation of the amount of intracellular polyP.
Detection of intracellular ATP in the ΔphoU1 and ΔphoU2 mutants.
The intracellular ATP levels in the ΔphoU1 and ΔphoU2 mutants were detected using an ATP colorimetric/fluorometric assay kit (catalog no. MAK190) from Sigma with modification for S. epidermidis. At the time points indicated above and in the relevant figures (when the OD600 was 0.5), the cells were pelleted by centrifugation at 4,000 × g at 4°C, washed with cold sterile PBS, resuspended in 1 ml of ATP extraction buffer within 10 min of initial pelleting of the culture, and lysed with 0.1 mm glass-silica beads in a BeadBeater apparatus (BioSpec). The resulting supernatant, obtained by centrifuging the samples at 20,000 × g for 15 min at 4°C, was filtered through a 10-kDa-cutoff filter, as suggested by the assay manufacturer, to remove enzymes that could otherwise deplete ATP.
Quantification of intracellular NADP+/NADPH in the ΔphoU1 and ΔphoU2 mutants.
NADP+ and NADPH were quantified as described by Posada et al. using an NADPH/NADP+ kit from Sigma, which was modified for S. epidermidis (66). Because NADPH can be unstable, the cells were extracted, filtered, and assayed as quickly as possible, essentially as described above.
Sensitivity of the ΔphoU1 and ΔphoU2 mutants to H2O2 and SDS.
Overnight cultures of the S. epidermidis strains were diluted 1:200 in fresh TSB medium and incubated at 37°C for 3 h until an OD600 of 1 was reached. After 10-fold serial dilution, 5 μl of the aliquot was spotted onto a TSB agar plate containing 6 mM H2O2 or 0.006% SDS and incubated at 37°C overnight. The bacterial colonies on the plates were photographed and counted (5, 31).
MIC and MBC.
The MICs of the antibiotics against the five S. epidermidis strains were determined using 2-fold serial dilutions of the antibiotics in Mueller-Hinton (MH) broth (Oxoid, Basingstoke, UK) according to CLSI guidelines (67). A log-phase culture (6 h) was adjusted to a turbidity equivalent to that of a 0.5 McFarland standard (108 CFU/ml), inoculated into MH broth (1:200), and then incubated at 37°C for 16 to 20 h. The MIC endpoint was defined as the lowest concentration at which there was no visible growth in the tubes. The broth containing no drugs served as a control. The minimal bactericidal concentration (MBC) values were assessed by plating 100-μl samples from each negative culture tube (tubes with no visible bacterial growth) from the MIC assays onto blank MH broth agar plates. The MBC was the concentration at which a 99.9% reduction of the original inoculum was observed.
Determination of antibiotic-tolerant bacteria.
Different antibiotics were added to the stationary-phase cultures (16 h) of the S. epidermidis strains (final concentrations, levofloxacin at 50 mg/liter, vancomycin at 75 mg/liter, and amikacin at 50 mg/liter), and then the cultures were incubated at 37°C for 120 h without shaking. One milliliter of the bacterial culture was then washed twice with ice-cold saline and pelleted by centrifugation for 3 min at 6,000 × g, and the pellet was resuspended in 1 ml cold saline, which was used to generate a series of 10-fold dilutions. Triplicate 5-μl aliquots from each dilution were spotted onto TSA plates for determination of the numbers of CFU (5). Three independent experiments were performed.
RNA isolation and RNA sequencing.
Total RNA was isolated from strain SE1457 and the ΔphoU1 and ΔphoU2 mutants using an RNeasy minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Briefly, bacterial cultures were centrifuged at 5,000 × g for 5 min, and then the pellets were washed twice in 0.9% saline. The culture was homogenized 5 times using 0.1-mm zirconia-silica beads in a mini-BeadBeater (Biospec, Bartlesville, OK) at 4,800 rpm for 40 s at 1-min intervals on ice. The RNA extracted using the silica-based filter was purified with phenol-chloroform-isoamyl alcohol and precipitated with absolute ethanol.
RNA-Seq was performed according to the Illumina RNA sequencing sample preparation guide, using three biological replicates for each of the S. epidermidis strains. Samples of SE1457 and ΔphoU1 and ΔphoU2 mutant RNA were treated with RNase-free DNase I (TaKaRa) to prevent contamination with genomic DNA. The RNA quality was evaluated using a Bioanalyzer 2100 system (Agilent Technologies Deutschland GmbH). Prior to the sequencing analysis, rRNA was removed with a RiboZero rRNA removal kit for Gram-positive organisms. After depletion of the rRNA, fragmented RNA was used as a template for PCR with random primers. The cDNA libraries were prepared by using an mRNA-Seq sample preparation kit (Illumina). The concentration of cDNA was measured using a Qubit (version 2.0) fluorometer, the fragment size (200 to 300 bp) was verified on a Bioanalyzer 2100 system, amplification was performed using an Illumina cBot system, and sequencing was performed with an Illumina HiSeq 2500 sequencer for 51 cycles; all of these procedures were performed according to the manufacturers' protocols. Raw sequencing data were processed using the data collection software provided by Illumina.
RNA-Seq data analysis and DEG validation by RT-qPCR.
Raw sequencing reads were preprocessed by filtering out rRNA reads, sequencing adapters, short fragment reads, and other low-quality reads. The remaining reads were mapped to the reference genome of S. epidermidis RP62A at the NCBI website with Bowtie2 software (version 2.0.5) on the basis of the local alignment algorithm. Reads that aligned to multiple locations were kept (to a maximum of 20 potential positions) to assist with the construction of gene models for genes with repetitive or low-complexity features. When the reads were aligned, 1 mismatch with the reference sequence was allowed. The alignments reported using Bowtie2 software were further processed with BEDTools software to determine the transcript expression levels and their differential expression between each two of the three samples. Expression values were presented as the number of reads per kilobase of genes per million mapped reads (RPKM). Data were visualized using the Integrated Genomics Viewer. Differential expression of all the transcripts was quantified using DEGseq software (version 2.16.1), and then the fold change values were presented. In general, we recorded no distinct difference between two transcripts when their fold change value fell between 0.666 and 1.5. Concomitantly, P values were determined, and significance was assessed by correcting for multiple testing using, for example, Fisher's exact test and the Wilcoxon test. Differentially expressed genes with corresponding expression values were uploaded into IPA software and analyzed using the canonical pathway. We used Fisher's exact test to select significant pathways, and the significance threshold was defined by a P value of 0.05. The significance of the pathway was indicated by the ratio of the number of genes from the data set that mapped to the pathway versus the total number of genes present in the canonical pathway.
For validation of DEGs identified by RNA-Seq, total RNA extracted from SE1457 and the ΔphoU1 and ΔphoU2 mutants was treated with the reagents from a PrimeScript RT reagent kit (TaKaRa Biotechnology, Dalian, China) for DNA digestion and reverse transcribed into cDNA. qPCRs were performed using a Mastercycler RealPlex system (Eppendorf AG, Hamburg, Germany) with SYBR green PCR reagents (SYBR Premix Ex Taq; TaKaRa Biotechnology, Dalian, China). The amplification conditions were 95°C for 30 s and 40 cycles of 95°C for 5 s and 60°C for 34 s, followed by a melting curve analysis. gyrB (DNA gyrase subunit B gene) was used as a housekeeping gene to normalize the transcript levels of the genes in the qPCR. All RT-qPCRs were performed in triplicate.
Construction of a protein-protein interaction network.
A protein-protein interaction network was constructed using Cytoscape software. A total of 174 DEGs encoding the candidate proteins involved in bacterial metabolism were extracted. The protein-protein interaction network was constructed according to the functional relationships annotated in the KEGG database (http://www.genome.jp/kegg/pathway.html). In the network, lines were used to represent protein-protein interactions, including binding/association, phosphorylation, activation, and inhibition. Proteins encoded by upregulated and downregulated DEGs are indicated in the figures in red and green, respectively.
Statistical analysis.
Experimental data were analyzed with SPSS software and compared using the Student t test or one-way analysis of variance. Differences with a P value of <0.05 were considered statistically significant.
Accession number(s).
The complete RNA-Seq data set is posted in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE97400 and GSE97656 for the original data set.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Zonghou Shen (Department of Biochemistry and Molecular Biology, Fudan University) for suggestions on analyzing the DGEs associated with major metabolic pathways.
This work was supported by the National Natural Science Foundation of China (81271791, 81571955, and 81573268) and the National High-Tech and Development Plan of China (2014AA021404).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00219-17.
REFERENCES
- 1.Götz F, Peters G. 2000. Colonization of medical devices by coagulase-negative staphylococci, p 55–88. In Waldvogel FA, Bisno AL (ed), Infections associated with indwelling medical devices, 3rd ed American Society for Microbiology, Washington, DC. [Google Scholar]
- 2.Maduka-Ezeh AN, Greenwood-Quaintance KE, Karau MJ, Berbari EF, Osmon DR, Hanssen AD, Steckelberg JM, Patel R. 2012. Antimicrobial susceptibility and biofilm formation of Staphylococcus epidermidis small colony variants associated with prosthetic joint infection. Diagn Microbiol Infect Dis 74:224–229. doi: 10.1016/j.diagmicrobio.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 3.Dunne WM. 2002. Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev 15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rupp ME, Fey PD, Heilmann C, Gotz F. 2001. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J Infect Dis 183:1038–1042. doi: 10.1086/319279. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Zhang Y. 2007. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob Agents Chemother 51:2092–2099. doi: 10.1128/AAC.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi W, Zhang Y. 2010. PhoY2 but not PhoY1 is the PhoU homologue involved in persisters in Mycobacterium tuberculosis. J Antimicrob Chemother 65:1237–1242. doi: 10.1093/jac/dkq103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, Mao Y, Yu J, Zhu L, Li M, Wang D, Dong D, Liu J, Gao Q. 2013. PhoY2 of mycobacteria is required for metabolic homeostasis and stress response. J Bacteriol 195:243–252. doi: 10.1128/JB.01556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Almeida LG, Ortiz JH, Schneider RP, Spira B. 2015. phoU inactivation in Pseudomonas aeruginosa enhances accumulation of ppGpp and polyphosphate. Appl Environ Microbiol 81:3006–3015. doi: 10.1128/AEM.04168-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Lou Y, Yokota H, Adams PD, Kim R, Kim SH. 2005. Crystal structure of a PhoU protein homologue: a new class of metalloprotein containing multinuclear iron clusters. J Biol Chem 280:15960–15966. doi: 10.1074/jbc.M414117200. [DOI] [PubMed] [Google Scholar]
- 10.Chan FY, Torriani A. 1996. PstB protein of the phosphate-specific transport system of Escherichia coli is an ATPase. J Bacteriol 178:3974–3977. doi: 10.1128/jb.178.13.3974-3977.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mechler L, Herbig A, Paprotka K, Fraunholz M, Nieselt K, Bertram R. 2015. A novel point mutation promotes growth phase-dependent daptomycin tolerance in Staphylococcus aureus. Antimicrob Agents Chemother 59:5366–5376. doi: 10.1128/AAC.00643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mechler L, Bonetti EJ, Reichert S, Flotenmeyer M, Schrenzel J, Bertram R, Francois P, Gotz F. 2016. Daptomycin tolerance in the Staphylococcus aureus pitA6 mutant is due to upregulation of the dlt operon. Antimicrob Agents Chemother 60:2684–2691. doi: 10.1128/AAC.03022-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 14.Otto M. 2008. Staphylococcal biofilms. Curr Top Microbiol Immunol 322:207–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkler ME, Hoch JA. 2008. Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J Bacteriol 190:2645–2648. doi: 10.1128/JB.01682-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masalha M, Borovok I, Schreiber R, Aharonowitz Y, Cohen G. 2001. Analysis of transcription of the Staphylococcus aureus aerobic class Ib and anaerobic class III ribonucleotide reductase genes in response to oxygen. J Bacteriol 183:7260–7272. doi: 10.1128/JB.183.24.7260-7272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs S, Pane-Farre J, Kohler C, Hecker M, Engelmann S. 2007. Anaerobic gene expression in Staphylococcus aureus. J Bacteriol 189:4275–4289. doi: 10.1128/JB.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leibig M, Liebeke M, Mader D, Lalk M, Peschel A, Gotz F. 2011. Pyruvate formate lyase acts as a formate supplier for metabolic processes during anaerobiosis in Staphylococcus aureus. J Bacteriol 193:952–962. doi: 10.1128/JB.01161-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y, Wu Y, Zhu T, Han H, Liu H, Xu T, Francois P, Fischer A, Bai L, Gotz F, Qu D. 2015. Staphylococcus epidermidis SrrAB regulates bacterial growth and biofilm formation differently under oxic and microaerobic conditions. J Bacteriol 197:459–476. doi: 10.1128/JB.02231-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazmierczak MJ, Wiedmann M, Boor KJ. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol Mol Biol Rev 69:527–543. doi: 10.1128/MMBR.69.4.527-543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heilmann C, Hussain M, Peters G, Gotz F. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol 24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 22.Lang S, Xu J, Stuart F, Thomas RM, Vrijbloed JW, Robinson JA. 2000. Analysis of antibody A6 binding to the extracellular interferon gamma receptor alpha-chain by alanine-scanning mutagenesis and random mutagenesis with phage display. Biochemistry 39:15674–15685. doi: 10.1021/bi000838z. [DOI] [PubMed] [Google Scholar]
- 23.Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 24.Hensley K, Robinson KA, Gabbita SP, Salsman S, Floyd RA. 2000. Reactive oxygen species, cell signaling, and cell injury. Free Radic Biol Med 28:1456–1462. doi: 10.1016/S0891-5849(00)00252-5. [DOI] [PubMed] [Google Scholar]
- 25.Nordberg J, Arner ES. 2001. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic Biol Med 31:1287–1312. doi: 10.1016/S0891-5849(01)00724-9. [DOI] [PubMed] [Google Scholar]
- 26.Lee SM, Koh HJ, Park DC, Song BJ, Huh TL, Park JW. 2002. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med 32:1185–1196. doi: 10.1016/S0891-5849(02)00815-8. [DOI] [PubMed] [Google Scholar]
- 27.Maeng O, Kim YC, Shin HJ, Lee JO, Huh TL, Kang KI, Kim YS, Paik SG, Lee H. 2004. Cytosolic NADP(+)-dependent isocitrate dehydrogenase protects macrophages from LPS-induced nitric oxide and reactive oxygen species. Biochem Biophys Res Commun 317:558–564. doi: 10.1016/j.bbrc.2004.03.075. [DOI] [PubMed] [Google Scholar]
- 28.Giro M, Carrillo N, Krapp AR. 2006. Glucose-6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli. Microbiology 152:1119–1128. doi: 10.1099/mic.0.28612-0. [DOI] [PubMed] [Google Scholar]
- 29.Marino D, Gonzalez EM, Frendo P, Puppo A, Arrese-Igor C. 2007. NADPH recycling systems in oxidative stressed pea nodules: a key role for the NADP+-dependent isocitrate dehydrogenase. Planta 225:413–421. [DOI] [PubMed] [Google Scholar]
- 30.Kornberg A. 1999. Inorganic polyphosphate: a molecule of many functions. Prog Mol Subcell Biol 23:1–18. doi: 10.1007/978-3-642-58444-2_1. [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Sun X, Wu Y, Xie C, Zhang W, Wang D, Chen X, Qu D, Gan J, Chen H, Jiang H, Lan L, Yang CG. 2013. Oxidation-sensing regulator AbfR regulates oxidative stress responses, bacterial aggregation, and biofilm formation in Staphylococcus epidermidis. J Biol Chem 288:3739–3752. doi: 10.1074/jbc.M112.426205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Surin BP, Rosenberg H, Cox GB. 1985. Phosphate-specific transport system of Escherichia coli: nucleotide sequence and gene-polypeptide relationships. J Bacteriol 161:189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SK, Makino K, Amemura M, Shinagawa H, Nakata A. 1993. Molecular analysis of the phoH gene, belonging to the phosphate regulon in Escherichia coli. J Bacteriol 175:1316–1324. doi: 10.1128/jb.175.5.1316-1324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morohoshi T, Maruo T, Shirai Y, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kuroda A. 2002. Accumulation of inorganic polyphosphate in phoU mutants of Escherichia coli and Synechocystis sp. strain PCC6803. Appl Environ Microbiol 68:4107–4110. doi: 10.1128/AEM.68.8.4107-4110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beard SJ, Hashim R, Wu G, Binet MR, Hughes MN, Poole RK. 2000. Evidence for the transport of zinc(II) ions via the pit inorganic phosphate transport system in Escherichia coli. FEMS Microbiol Lett 184:231–235. doi: 10.1111/j.1574-6968.2000.tb09019.x. [DOI] [PubMed] [Google Scholar]
- 36.Vershinina OA, Znamenskaia LV. 2002. The Pho regulons of bacteria. Mikrobiologiia 71:581–595. (In Russian.) [PubMed] [Google Scholar]
- 37.Steed PM, Wanner BL. 1993. Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J Bacteriol 175:6797–6809. doi: 10.1128/jb.175.21.6797-6809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun G, Sharkova E, Chesnut R, Birkey S, Duggan MF, Sorokin A, Pujic P, Ehrlich SD, Hulett FM. 1996. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J Bacteriol 178:1374–1385. doi: 10.1128/jb.178.5.1374-1385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roca I, Torrents E, Sahlin M, Gibert I, Sjoberg BM. 2008. NrdI essentiality for class Ib ribonucleotide reduction in Streptococcus pyogenes. J Bacteriol 190:4849–4858. doi: 10.1128/JB.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinkel TL, Roux CM, Dunman PM, Fang FC. 2013. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio 4:e00696-13. doi: 10.1128/mBio.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabret C, Hoch JA. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol 180:6375–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dubrac S, Bisicchia P, Devine KM, Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol 70:1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- 43.Chai Y, Beauregard PB, Vlamakis H, Losick R, Kolter R. 2012. Galactose metabolism plays a crucial role in biofilm formation by Bacillus subtilis. mBio 3:e00184-12. doi: 10.1128/mBio.00184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bera A, Biswas R, Herbert S, Kulauzovic E, Weidenmaier C, Peschel A, Gotz F. 2007. Influence of wall teichoic acid on lysozyme resistance in Staphylococcus aureus. J Bacteriol 189:280–283. doi: 10.1128/JB.01221-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlag M, Biswas R, Krismer B, Kohler T, Zoll S, Yu W, Schwarz H, Peschel A, Gotz F. 2010. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol 75:864–873. doi: 10.1111/j.1365-2958.2009.07007.x. [DOI] [PubMed] [Google Scholar]
- 46.Andrey DO, Renzoni A, Monod A, Lew DP, Cheung AL, Kelley WL. 2010. Control of the Staphylococcus aureus toxic shock tst promoter by the global regulator SarA. J Bacteriol 192:6077–6085. doi: 10.1128/JB.00146-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao NN, Gomez-Garcia MR, Kornberg A. 2009. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem 78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- 48.Davidson AL, Dassa E, Orelle C, Chen J. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K. 2016. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol 1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 50.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 52.Zhu T, Lou Q, Wu Y, Hu J, Yu F, Qu D. 2010. Impact of the Staphylococcus epidermidis LytSR two-component regulatory system on murein hydrolase activity, pyruvate utilization and global transcriptional profile. BMC Microbiol 10:287. doi: 10.1186/1471-2180-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lou Q, Zhu T, Hu J, Ben H, Yang J, Yu F, Liu J, Wu Y, Fischer A, Francois P, Schrenzel J, Qu D. 2011. Role of the SaeRS two-component regulatory system in Staphylococcus epidermidis autolysis and biofilm formation. BMC Microbiol 11:146. doi: 10.1186/1471-2180-11-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Helle L, Kull M, Mayer S, Marincola G, Zelder ME, Goerke C, Wolz C, Bertram R. 2011. Vectors for improved Tet repressor-dependent gradual gene induction or silencing in Staphylococcus aureus. Microbiology 157:3314–3323. doi: 10.1099/mic.0.052548-0. [DOI] [PubMed] [Google Scholar]
- 55.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl Environ Microbiol 70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gust AA, Biswas R, Lenz HD, Rauhut T, Ranf S, Kemmerling B, Gotz F, Glawischnig E, Lee J, Felix G, Nurnberger T. 2007. Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J Biol Chem 282:32338–32348. doi: 10.1074/jbc.M704886200. [DOI] [PubMed] [Google Scholar]
- 57.Vollmer W, Seligman SJ. 2010. Architecture of peptidoglycan: more data and more models. Trends Microbiol 18:59–66. doi: 10.1016/j.tim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 58.Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol 22:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 60.Hu J, Xu T, Zhu T, Lou Q, Wang X, Wu Y, Huang R, Liu J, Liu H, Yu F, Ding B, Huang Y, Tong W, Qu D. 2011. Monoclonal antibodies against accumulation-associated protein affect EPS biosynthesis and enhance bacterial accumulation of Staphylococcus epidermidis. PLoS One 6:e20918. doi: 10.1371/journal.pone.0020918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lam H, Kesselly A, Stegalkina S, Charlebois RL, Oomen R, Kleanthous H, Yethon JA. 2015. Correction for Lam et al., antibodies to PhnD inhibit staphylococcal biofilms. Infect Immun 83:2197. doi: 10.1128/IAI.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brunskill EW, Bayles KW. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol 178:611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gerke C, Kraft A, Sussmuth R, Schweitzer O, Gotz F. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem 273:18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 64.Wu Y, Wang J, Xu T, Liu J, Yu W, Lou Q, Zhu T, He N, Ben H, Hu J, Gotz F, Qu D. 2012. The two-component signal transduction system ArlRS regulates Staphylococcus epidermidis biofilm formation in an ica-dependent manner. PLoS One 7:e40041. doi: 10.1371/journal.pone.0040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aschar-Sobbi R, Abramov AY, Diao C, Kargacin ME, Kargacin GJ, French RJ, Pavlov E. 2008. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J Fluoresc 18:859–866. doi: 10.1007/s10895-008-0315-4. [DOI] [PubMed] [Google Scholar]
- 66.Posada AC, Kolar SL, Dusi RG, Francois P, Roberts AA, Hamilton CJ, Liu GY, Cheung A. 2014. Importance of bacillithiol in the oxidative stress response of Staphylococcus aureus. Infect Immun 82:316–332. doi: 10.1128/IAI.01074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turnidge J, Bordash G. 2007. Statistical methods for establishing quality control ranges for antibacterial agents in Clinical and Laboratory Standards Institute susceptibility testing. Antimicrob Agents Chemother 51:2483–2488. doi: 10.1128/AAC.01457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.