ABSTRACT
The complete nucleotide sequences of six IMP-4-encoding plasmids recovered from Enterobacteriaceae isolates of wildlife origin were characterized. Sequencing data showed that plasmids of different incompatibility groups (IncM, IncI1, IncF, and nontypeable [including an IncX5_2 and two pPrY2001-like]) carried the blaIMP-4-carrying integrons In809 or In1460. Most of the plasmids carried an mph(A) region, and chrA-like, aac(3)-IId, and blaTEM-1b genes. Finally, plasmid analysis revealed the involvement of two different IS26- and Tn1696-associated mechanisms in the mobilization of IMP-4-encoding integrons.
KEYWORDS: IncM, IncI1, IncF, IncX5_2, Enterobacter aerogenes, Escherichia coli, Proteus mirabilis, Proteus penneri, In809, In1460
TEXT
Acquired carbapenem-hydrolyzing metallo-β-lactamases (MBLs) are resistance determinants in Gram-negative pathogens that are of increasing clinical importance. Of these, IMP-4 may be the most prevalent carbapenemase in certain regions (1, 2), but globally, NDM-type MBLs are more frequently reported. In a previous study from our group, a total of 116 IMP-4-producing Enterobacteriaceae isolates were identified in cloacal samples that were collected from 80 out of 200 (40%) gull chicks at a nesting colony in New South Wales, Australia (3). The blaIMP-4 gene was carried by plasmids of variable sizes and by diverse replicons, including HI2-N (n = 30), HI2 (n = 11), A/C (n = 17), A/C-Y (n = 2), L/M (n = 5), I1 (n = 1), and nontypeable (n = 6). However, the complete nucleotide sequences of blaIMP-4-carrying plasmids, being positive for HI2 or A/C replicons, have been previously described (4, 5). Therefore, in the present study, we characterized the complete nucleotide sequences of blaIMP-4-carrying plasmids, which were assigned to different incompatibility (Inc) groups using a PCR-based replicon typing (PBRT) method (6), in order to examine the nature of the genetic mobile elements involved in the acquisition and spread of blaIMP-4 in Enterobacteriaceae of wildlife origin.
Six blaIMP-4-carrying plasmids representative of different sizes and replicons (Table 1) were selected for complete sequencing. Five of the blaIMP-4-carrying plasmids were transferred by conjugation from clinical strains, although with different transfer frequencies (Table 1), while the remaining plasmid was transferred by transformation. All IMP-4-producing recombinants exhibited resistance to cefotaxime and ceftazidime (see Table S1 in the supplemental material), while they remained susceptible to piperacillin-tazobactam and carbapenems. Variations observed in the MICs of piperacillin and aztreonam might reflect the presence of additional resistance mechanisms in some of the recombinants. Plasmids were extracted from Escherichia coli transconjugants or transformants and were sequenced using the Illumina MiSeq platform (Illumina Inc., San Diego, CA). Initial paired-end reads were quality trimmed using the Trimmomatic tool v0.36 (7) with a sliding window size of 4 bp, required average base quality of ≥17, and minimum read length of 48 bases. For assembly of the plasmids, reads were mapped to the reference E. coli strain K-12 substrain MG1655 genome (GenBank accession no. U00096) using the BWA-MEM algorithm (8), in order to filter out the chromosomal DNA. All of the unmapped reads were then assembled by use of the de Bruijn graph-based de novo assembler SPAdes v3.11.0 (9). The sequence gaps were filled by a PCR-based strategy and Sanger sequencing.
TABLE 1.
Characteristics of IMP-4-encoding plasmids recovered from Enterobacteriaceae of wildlife origin
| Plasmid | Source strain | Size of blaIMP-4-positive plasmid (kb)a | PBRTb | Replicon(s) | Conjugal transfer frequencyc | Integron with blaIMP-4 gene | β-Lactamase content | Additional resistance gene(s) |
|---|---|---|---|---|---|---|---|---|
| pEa1631 | Enterobacter aerogenes | 85.489 | L/M | IncM | 5 × 10−2 | In809 | IMP-4, TEM-1 | aac(3)-IId, mph(A) |
| pEc42 | E. coli ST695 | 112.530 | I1 | IncI1 | 5 × 10−5 | In809 | IMP-4 | mph(A), qnrB2 |
| pEc1675 | E. coli ST542 | 158.433 | NT | IncFII, IncFIB(K), plasmid B-like repA | ND | In809 | IMP-4, SHV-12, OXA-1, TEM-1 | aac(3)-IId, mph(A), arr-3, dfrA19 |
| pPp47 | Proteus penneri | 142.085 | NT | pPrY2001-like repB | 6 × 10−4 | In1460 | IMP-4 | aac(3)-IId, mph(A), aphA1, tet(B) |
| pPm60 | Proteus mirabilis | 113.297 | NT | pPrY2001-like repB | 2 × 10−2 | In809 | IMP-4 | aac(3)-IId, dfrA1, sat2, aadA1a, qacH2, sul3, aphA1, tet(B), lnu(G) |
| pEc1677 | E. coli ST1139 | 69.785 | NT | IncX5_2 | 3 × 10−7 | In809 | IMP-4 | mph(A) |
Data for plasmids found in transconjugants are shown in boldface type; datum for plasmids observed in transformants is underlined.
PBRT, PCR-based replicon typing; NT, nontypeable.
Transfer frequencies are expressed as transconjugants per donor cell. ND, not determined.
Plasmid pEc42 was also sequenced using MinION technology, which provides long reads of single-molecule DNA and enables the closing of the whole plasmid sequence in repetitive shufflon regions. Library preparations were performed using a SQK-LSK208 ligation sequencing 2D kit according to the manufacturer's protocol. The library was loaded onto a flow cell (FLO-MIN106 R9.4 SpotON) and sequenced for 48 h. Base calling was performed in real time via ONT's Metrichor service (desktop agent v2.43.1 and 2D basecalling workflow v1.125). After passing Metrichor quality control, MinION 2D reads were converted from fast5 to fastq format using the poretools toolkit (10). Short Illumina reads were trimmed using Trimmomatic (7). MinION reads yielded 2,937,908,973 bases, with a read count of 442,546 reads (N50 = 14,235 bp). The long reads from MinION were used as a scaffold, and along with short Illumina reads they were assembled using SPAdes hybrid assembly (9). Hybrid assembly produced a unique contig of 112,530 bp, with a k-mer coverage of 565×.
For sequence analysis and annotation, the BLAST algorithm (www.ncbi.nlm.nih.gov/BLAST), the ISfinder database (www-is.biotoul.fr/), and the ORF Finder tool (www.bioinformatics.org/sms/) were utilized. Comparative genome alignments were performed using the Mauve v2.3.1 program (11).
Sequence analysis demonstrated that, in all plasmids (except pPp47), the blaIMP-4 gene occurred as the first cassette of the class 1 integron In809, which also included an array of qacG2, aacA4, and catB3 gene cassettes. Plasmid pPp47 had a new integron, In1460, whose variable region comprised the blaIMP-4, qacG2, aacA4, and qacG cassettes. The variable region of In1460 was interrupted by insertion of a group IIc intron, downstream of the attC site of the aacA4 gene cassette.
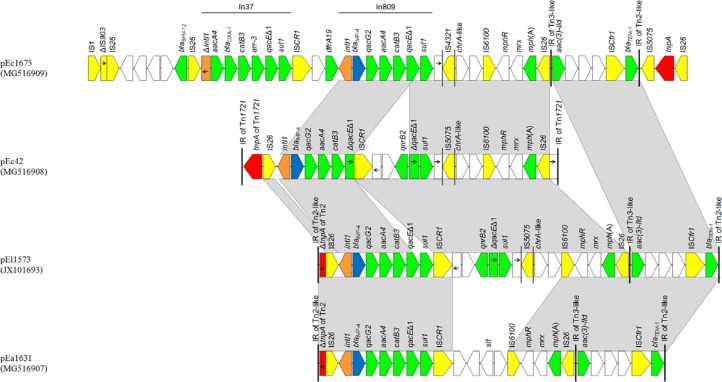
Plasmid pEa1631, being an 85,489-bp molecule, is a derivative of the IncM blaIMP-4-carrying plasmid pEl1573 (94% coverage and 100% identity) (see Fig. S1 in the supplemental material) described from a clinical isolate of Enterobacter cloacae (El1573) recovered in Sydney in 2004 (12). Plasmid pEa1631 was composed of a contiguous segment of 60,176 bp (nt 1 to 53,777 and 79,091 to 85,489) sharing extensive similarity with the backbone of IncM plasmids (12, 13) and a 25,313-bp multidrug resistance (MDR) sequence (nt 53,778 to 79,090) inserted in the same position as that reported in pEl1573. Plasmid pEa1631 differed from pEl1573 by the deletion of a 6,920-bp segment, including the qnrB2 gene, a second copy of an integron 3′ conserved sequence (3′-CS), IS5075, and chrA-like (Fig. 1). The deleted segment was occupied by a 4,678-bp sequence (nt 62,349 to 67,026 in pEa1631), which exhibited 99% identity with a contiguous region described in the chromosomes of Klebsiella pneumoniae isolates, like K. pneumoniae ATCC 35657 (GenBank accession no. CP015134). A similar sequence has also been observed in the A/C2 plasmids pCf52 and pCf53, which were characterized from the same collection of IMP-producing Enterobacteriaceae isolates (5).
FIG 1.
Detailed comparison of the MDR regions of the plasmids pEa1631, pEc42, pEc1675, and pEl1573 (12). Arrows show the direction of transcription of open reading frames (ORFs), while truncated ORFs appear as rectangles (arrows within rectangles indicate the direction of transcription). Resistance genes, IS elements, and transposases are shown in green, yellow, and red, respectively. blaIMP-4 genes are shown in blue, while intI1 genes are shaded orange. The remaining genes are shown in white. Inverted repeats (IRs) of transposon-derived sequences are indicated by vertical lines. Homologous segments (representing ≥99% sequence identity) are indicated by light gray shading.
Plasmid pEc42 is a 112,530-bp molecule with a sequence closely related to those of the IncI1 plasmids, like pEC008 (88% coverage and 99% identity) (Fig. S1) from an E. coli strain EC008 isolate recovered from a chicken (GenBank accession no. KY748190). Analysis by the pMLST Web tool (14) indicated that pEc42 belonged to sequence type (ST) 136. Plasmid pEc42 included an 88,254-bp IncI1 backbone (nt 1 to 3,928 and 28,205 to 112,530) and an MDR region of 24,276 bp (nt 3929 to 28204). The MDR region of pEc42, which exhibited a high similarity score with the respective region of pEl1573 (79% coverage and 99% identity) (12), carried the class 1 integron In809, qnrB2 and chrA-like genes, and an mph(A) region, and was bracketed by two IS26 copies in parallel orientation (Fig. 1). The MDR region of pEc42 was bounded by the two outer ends of Tn1721 and was inserted in the same position as Tn1721 in pEC008.
The non-self-transferable plasmid pEc1675 is 158,433 bp in size, and sequence analysis demonstrated that it is an IncF multireplicon molecule. The plasmid showed a complex structure, being composed of sequences of diverse origin and punctuated by mobile elements. A 51,871-bp sequence (nt 1 to 16,490, 38,675 to 53,602, 60,765 to 73,068, and 150,732 to 158,433) resembled that of the backbone of plasmid B (GenBank accession no. CP010244) (Fig. S1), including regions responsible for replication (IncFIB replicon [repA1 gene]) and plasmid maintenance (parB gene). Additionally, pEc1675 included a 28,112-bp sequence (nt 79,096 to 99,469 and 142,174 to 149,911) sharing common features with the backbone of the plasmid pABWA45_1 (GenBank accession no. CP022155). This sequence was composed of the IncFIB(K) (repB gene) and IncFII (repA2 gene) replicons and the maintenance sopAB and pemIK operons. The 42,704-bp (nt 99,470 to 142,173) MDR region of pEc1675, which exhibited extensive similarity with the respective region of pEl1573 (83% coverage and 99% identity) (12), contained the IMP-4-encoding integron In809, chrA-like, aac(3)-IId and blaTEM-1b genes, and an mph(A) region (Fig. 1). Furthermore, an IS26-bounded sequence encoding an SHV-12 extended-spectrum β-lactamase (ESBL) (15) and an integron similar to In37 from pHSH2, whose variable region comprised the aacA4, blaOXA-1, catB3, and arr-3 cassettes (16), were identified in the MDR region of pEc1675. Of note, a 213-bp segment of In37, including the 5′ end of the intI1 gene and the attI1 site, was deleted. The 3′ conserved sequence (CS) of In37 was disrupted by a copy of ISCR1, followed by the dfrA19 resistance gene. The MDR region of pEc1675 was bracketed by intact copies of IS1 and IS26 elements. No resistance genes were found in the three remaining acquired sequences, which carried intact and truncated copies of several mobile elements (IS1 [n = 3], IS26 [n = 3], IS102-like, IS903B, ΔTn3-like, ΔTn1000-like, ΔISKpn26, ΔISEc25, and ΔIS1) and a sequence of Alteromonas sp. origin (GenBank accession no. CP018023).
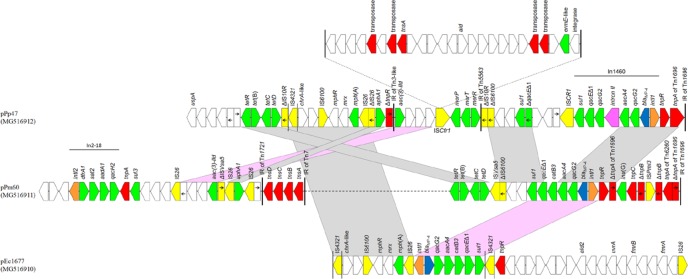
Plasmid pPp47, which was nontypeable by the PBRT method (6), was composed of two distinct parts: a contiguous plasmid backbone of 71,893 bp (nt 1 to 69,609 and 139,802 to 142,085) and an acquired sequence of 70,192 bp (nt 69,610 to 139,801). The plasmid backbone, which shared similarities with the respective regions of nontypeable NDM-1-encoding plasmids recovered from Providencia rettgeri from Canada and Latin America (17, 18), harbored coding sequences for replication (repB gene), conjugative transfer (tra genes), and plasmid maintenance (parAG and mazEF operons, and umuC, ssb, and parB genes) (Fig. S1). The acquired sequence of pPp47 contained In1460, located in the Tn1696 module (Fig. 2), in precisely the same position as that of In4 in Tn1696. Interestingly, IMP-encoding In809-like integrons associated with Tn1696-like transposons were identified in A/C2 plasmids characterized from Enterobacteriaceae isolates from the same collection (5). In addition to an mph(A) region and chrA-like and aac(3)-IId genes, the acquired sequence of pPp47 contained parts of transposons Tn5563α with a mercury resistance operon (mer) (19), Tn10 including genes for tetracycline resistance (tet genes) (20), and Tn4352-like with the kanamycin resistance gene aphA1. Additionally, a 28,513-bp segment (nt 90,295 to 118,807) was inserted into a copy of an ISCfr1 element. This segment, which was flanked by 28-bp inverted repeats (IR), harbored coding sequences for putative transposases, alcohol dehydrogenases, and proteins with unknown functions. Parts of this segment have been previously found in the chromosome I of Vibrio anguillarum strain VIB43 (GenBank accession no. CP023054) and in the pPrY2001-like CTX-M-131-encoding plasmid pC131 (GenBank accession no. KX774387) from P. rettgeri strain 30905. Target site duplications of 5 bp (CCTAG) at the boundaries of the latter segment indicated insertion by transposition.
FIG 2.
Detailed comparison of the MDR regions of the plasmids pPp47, pPm60, and pEc1677. Arrows show the direction of transcription of ORFs, while truncated ORFs appear as rectangles (arrows within rectangles indicate the direction of transcription). Resistance genes, IS elements, and transposases are shown in green, yellow, and red, respectively. blaIMP-4 genes are shown in blue, while intI1 and intI2 genes are shaded orange. A purple arrow indicates the group II intron; the remaining genes are shown in white. Inverted repeats (IRs) of transposon-derived sequences are indicated by vertical lines. Homologous segments (representing ≥99% sequence identity) are indicated by light gray shading, while pink shading shows inverted homologous segments.
The nontypeable plasmid pPm60 is a 113,297-bp molecule that is a derivative of pPrY2001-like plasmids (17, 18). pPm60 was composed of a pPrY2001-like backbone of 68,194 bp (nt 1 to 56,349, 80,589 to 90,149, and 111,014 to 113,297) and two acquired sequences (Fig. S1). The IMP-4-encoding acquired sequence (nt 90,150 to 111,013) was inserted in exactly the same position as that in pPp47. This sequence contained In809, with the IRi of the integron located between resI and resII sites of the Tn1696 module (Fig. 2). However, a Tn6260-like transposon carrying a lnu(G) gene for resistance to lincomycin was inserted into tnpA gene of the Tn1696 module. Tn6260 was originally described in a swine Enterococcus faecalis isolate from China (21). Downstream from the 3′-CS of In809, a part of Tn10 with tet genes (20) was found. The second acquired sequence (nt 56,350 to 80,588) of pPm60 carried the class 2 integron In2-18, whose variable region comprised the dfrA1, sat2, aadA1a, and qacH2 cassettes (GenBank accession no. HQ386833). Furthermore, the latter sequence included the Tn4352-like composite transposon, with aphA1 bounded by two IS26 elements in parallel orientation, and aac(3)-IId and sul3 genes.
Finally, plasmid pEc1677, which was nontypeable by the PBRT method, (6) was a 69,785-bp molecule. pEc1677 comprised two distinct parts: a contiguous plasmid backbone of 33,685 bp (nt 1 to 29,590 and 65,691 to 69,785), sharing extensive similarity (100% coverage and 99% identity) with the respective region of plasmid pCAV1043-58 (GenBank accession no. CP011588) (Fig. S1), and an acquired sequence of 36,100 bp (nt 29,591 to 65,690) carrying In809. The plasmid backbone was composed of regions responsible for replication (repB gene), conjugative transfer (pilX1-11 genes) and plasmid maintenance (parB, topB, ardR, taxA, taxB, and taxC genes) (Fig. S1). The transfer region of pEc1677 possessed extensive structural as well as sequence similarity (94%) with the transfer region of the IncX5 plasmid pBK31567 (22). Additionally, the taxC gene of pEc1677 exhibited 95% similarity with that of pBK31567 (22, 23). Therefore, pEc1677 was designated IncX5_2. However, pEc1677 had a repB gene showing limited similarity (59% coverage and 69% identity) to the respective gene of the aforementioned plasmid. In the acquired region of pEc1677, an mph(A) region and aac(3)-IId gene were identified (Fig. 2) downstream from the 5′-CS of In809. The 3′-CS of In809 was disrupted by a Tn6317-like module (24) composed of a 702-bp sequence that included a gene encoding a DNA invertase and the 38-bp inverted repeat (IR) of the transposon, which was interrupted by IS4321. Next to the Tn6317-like module, a 19,206-bp segment (nt 45,666 to 64,870) harboring coding sequences for an alcohol dehydrogenase and ABC transporters was found. The Tn6317-like fragment and 19,206-bp segment have been previously found in A/C2 plasmids pEc9 and pEc19 from the same collection of IMP-4-producing Enterobacteriaceae isolates (5). Of note, part of the pEc1677 acquired region, including the IMP-4-encoding integron In809, was bracketed by two copies of IS26, thus forming a composite transposon that could be implicated in the mobilization of this region.
In conclusion, the present study reported the complete nucleotide sequences of six different plasmids recovered from wildlife, carrying the IMP-4-encoding integrons In809 and In1460. Even though common characteristics [chrA-like, aac(3)-IId and blaTEM-1b genes, and an mph(A) region] were observed in the MDR regions of the characterized plasmids, unique features (like the SHV-12-encoding region in pEc1675, segments of Vibrio origin in pPp47, and Tn6260 and In2-18 in pPm60) were found in most of the plasmid sequences, underscoring the high plasticity of MDR regions. Of note, plasmid analysis revealed the involvement of two different mechanisms in the mobilization of IMP-4-encoding integrons, namely, (i) in plasmids pEa1631, pEc42, pEc1675, and pEc1677, the In809-carrying MDR regions were bracketed by intact copies of IS26 or other insertion elements, while (ii) in the pPrY2001-like plasmids pPp47 and pPm60, the In809 and In1460 integrons were located in the Tn1696 module. Association of In809-like integrons with the Tn1696 module has been previously demonstrated in A/C2 plasmids from Enterobacteriaceae of wildlife origin (5). These findings underscored the significant role of mobile elements in the reshuffling of enterobacterial plasmids and mobilization of large MDR segments.
Accession number(s).
The nucleotide sequences of the plasmids pEa1631, pEc42, pEc1675, pEc1677, pPm60, and pPp47 have been deposited in GenBank under the accession numbers MG516907, MG516908, MG516909, MG516910, MG516911, and MG516912, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to Iva Kutilova, Katarina Murgasova, Eva Suchanova, and Jarmila Lausova for their assistance in the laboratory, and to Ivo Papousek for assistance with the data analysis. We also thank Thomas Jové for curating integron sequencing data.
This work was supported by the Czech Science Foundation (grant 15-14683Y) and the National Sustainability Programs I (NPU I; grant LO1503) and II (NPU II; grant LQ1601) provided by the Ministry of Education Youth and Sports of the Czech Republic. A.V. was supported by the Internal Grant Agency of Veterinary and Pharmaceutical University Brno (grant 226/2017/FVHE).
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02434-17.
REFERENCES
- 1.Leung GH, Gray TJ, Cheong EY, Haertsch P, Gottlieb T. 2013. Persistence of related bla-IMP-4 metallo-beta-lactamase producing Enterobacteriaceae from clinical and environmental specimens within a burns unit in Australia—a six-year retrospective study. Antimicrob Resist Infect Control 2:35. doi: 10.1186/2047-2994-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu L, Zhong Q, Shang Y, Wang H, Ning C, Li Y, Hang Y, Xiong J, Wang X, Xu Y, Qin Z, Parsons C, Wang L, Yu F. 2014. The prevalence of carbapenemase genes and plasmid-mediated quinolone resistance determinants in carbapenem-resistant Enterobacteriaceae from five teaching hospitals in central China. Epidemiol Infect 142:1972–1977. doi: 10.1017/S0950268813002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dolejska M, Masarikova M, Dobiasova H, Jamborova I, Karpiskova R, Havlicek M, Carlile N, Priddel D, Cizek A, Literak I. 2016. High prevalence of Salmonella and IMP-4-producing Enterobacteriaceae in the silver gull on Five Islands, Australia. J Antimicrob Chemother 71:63–70. doi: 10.1093/jac/dkv306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abraham S, O'Dea M, Trott DJ, Abraham RJ, Hughes D, Pang S, McKew G, Cheong EY, Merlino J, Saputra S, Malik R, Gottlieb T. 2016. Isolation and plasmid characterization of carbapenemase (IMP-4) producing Salmonella enterica Typhimurium from cats. Sci Rep 6:35527. doi: 10.1038/srep35527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papagiannitsis CC, Kutilova I, Medvecky M, Hrabak J, Dolejska M. 2017. Characterization of the complete nucleotide sequences of IncA/C2 plasmids carrying In809-like integrons from Enterobacteriaceae isolates of wildlife origin. Antimicrob Agents Chemother 61:e01093-17. doi: 10.1128/AAC.01093-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 7.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv arXiv:1303.3997 [q-bio.GN].
- 9.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prijibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loman NJ, Quinlan AR. 2014. Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics 30:3399–3401. doi: 10.1093/bioinformatics/btu555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Partridge SR, Ginn AN, Paulsen IT, Iredell JR. 2012. pEl1573 carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob Agents Chemother 56:6029–6032. doi: 10.1128/AAC.01189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carattoli A, Seiffert SN, Schwendener S, Perreten V, Endimiani A. 2015. Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS One 10:e0123063. doi: 10.1371/journal.pone.0123063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miriagou V, Carattoli A, Tzelepi E, Villa L, Tzouvelekis LS. 2005. IS26-associated In4-type integrons forming multiresistance loci in enterobacterial plasmids. Antimicrob Agents Chemother 49:3541–3543. doi: 10.1128/AAC.49.8.3541-3543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M, Tran JH, Jacoby GA, Zhang Y, Wang F, Hooper DC. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob Agents Chemother 47:2242–2248. doi: 10.1128/AAC.47.7.2242-2248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mataseje LF, Boyd DA, Lefebvre B, Bryce E, Embree J, Gravel D, Katz K, Kibsey P, Kuhn M, Langley J, Mitchell R, Roscoe D, Simor A, Taylor G, Thomas E, Turgeon N, Mulvey MR; Canadian Nosocomial Infection Surveillance Program. 2014. Complete sequences of a novel blaNDM-1-harbouring plasmid from Providencia rettgeri and an FII-type plasmid from Klebsiella pneumoniae identified in Canada. J Antimicrob Chemother 69:637–642. doi: 10.1093/jac/dkt445. [DOI] [PubMed] [Google Scholar]
- 18.Marquez-Ortiz RA, Haggerty L, Olarte N, Duarte C, Garza-Ramos U, Silva-Sanchez J, Castro BE, Sim EM, Beltran M, Moncada MV, Valderrama A, Castellanos JE, Charles IG, Vanegas N, Escobar-Perez J, Petty NK. 2017. Genomic epidemiology of NDM-1-encoding plasmids in Latin American clinical isolates reveals insights into the evolution of multidrug resistance. Genome Biol Evol 9:1725–1741. doi: 10.1093/gbe/evx115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szuplewska M, Ludwiczak M, Lyzwa K, Czarnecki J, Bartosik D. 2014. Mobility and generation of mosaic non-autonomous transposons by Tn3-derived inverted-repeat miniature elements (TIMEs). PLoS One 9:e105010. doi: 10.1371/journal.pone.0105010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawley TD, Burland V, Taylor DE. 2000. Analysis of the complete nucleotide sequence of the tetracycline-resistance transposon Tn10. Plasmid 43:235–239. doi: 10.1006/plas.1999.1458. [DOI] [PubMed] [Google Scholar]
- 21.Zhu XQ, Wang XM, Li H, Shang YH, Pan YS, Wu CM, Wang Y, Du XD, Shen JZ. 2017. Novel lnu(G) gene conferring resistance to lincomycin by nucleotidylation, located on Tn6260 from Enterococcus faecalis E531. J Antimicrob Chemother 72:993–997. doi: 10.1093/jac/dkw549. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob Agents Chemother 57:269–276. doi: 10.1128/AAC.01648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Sun F, Zhou D, Sun Q, Luo W, Tong Y, Zhang D, Wang Q, Feng W, Chen W, Fan Y, Xia P. 2016. Genetic characterization of two fully sequenced multi-drug resistant plasmids pP10164-2 and pP10164-3 from Leclercia adecarboxylata. Sci Rep 6:33982. doi: 10.1038/srep33982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.