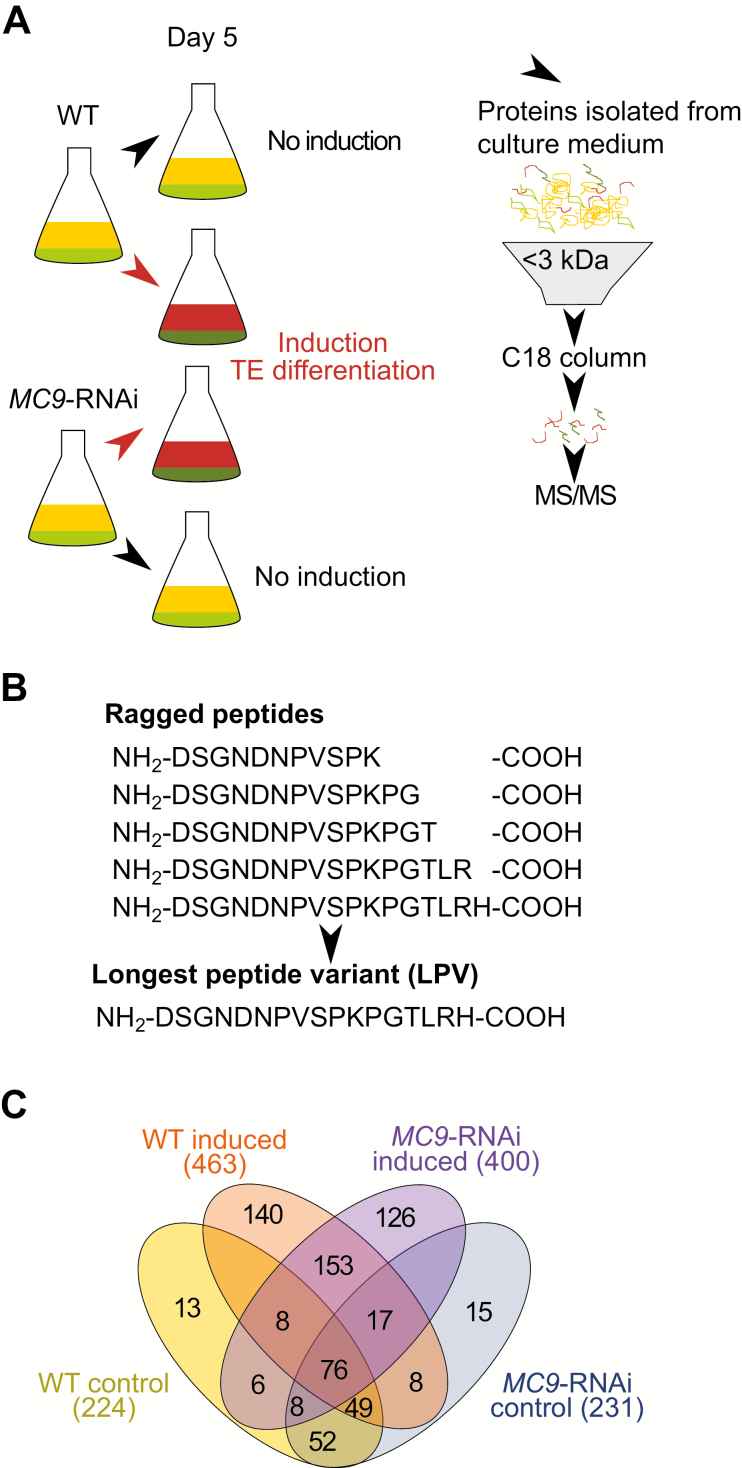
During development or stress response, plants need mechanisms to restrict cell death to the target cells. Kratos is the first plant extracellular peptide found to restrict cell death.
Keywords: Arabidopsis, autophagy, cell death, peptide, peptidomics, programmed cell death, stress response, vascular development, xylem
Abstract
During plant vascular development, xylem tracheary elements (TEs) form water-conducting, empty pipes by genetically regulated cell death. Cell death is prevented from spreading to non-TEs by unidentified intercellular mechanisms, downstream of METACASPASE9 (MC9)-mediated regulation of autophagy in TEs. Here, we identified differentially abundant extracellular peptides in vascular-differentiating wild-type and MC9-down-regulated Arabidopsis cell suspensions. A peptide named Kratos rescued the abnormally high ectopic non-TE death resulting from either MC9 knockout or TE-specific overexpression of the ATG5 autophagy protein during experimentally induced vascular differentiation in Arabidopsis cotyledons. Kratos also reduced cell death following mechanical damage and extracellular ROS production in Arabidopsis leaves. Stress-induced but not vascular non-TE cell death was enhanced by another identified peptide, named Bia. Bia is therefore reminiscent of several known plant cell death-inducing peptides acting as damage-associated molecular patterns. In contrast, Kratos plays a novel extracellular cell survival role in the context of development and during stress response.
Introduction
Genetically regulated cell death has important roles in stress responses, immunity, and development of multicellular organisms (Clarke and Clarke, 1996; Inohara and Nuñez, 2003; Galluzzi et al., 2015; Daneva et al., 2016; Huysmans et al., 2017). Upon stress, infection, or developmental stimuli, specific cells become committed to cell death and organized dismantlement. Such cell elimination events, unlike purely accidental cell death, rely on genetic regulation and occur in an anatomically organized manner often referred to as programmed cell death (PCD) (Lockshin and Williams, 1964; Lockshin and Zakeri, 2001; van Doorn et al., 2011; Galluzzi et al., 2015). In contrast to PCD, which implies a mechanism of cell suicide (Reynolds, 2014), cell death can also spread from target cells to neighbouring cells through non-cell autonomous mechanisms (Galluzzi et al., 2015; Escamez and Tuominen, 2017). In animals, a well-known example of such spreading cell death occurs during inflammation (Wallach et al., 2014; Kang et al., 2015; Zhong et al., 2016; Deretic and Levine, 2018). Generally, this latter type of ‘runaway’ cell death, here referred to as ‘ectopic’ cell death, is also genetically regulated even though the underlying molecular mechanisms in plants are poorly understood (Jabs et al., 1996; Endo et al., 2001; Mach et al., 2001; Brodersen et al., 2002; Tuominen et al., 2004; Overmyer et al., 2005; Wrzaczek et al., 2009, 2015; Coll et al., 2010; Escamez et al., 2016).
While inflammation in animals relies on the same core machinery in different situations (Wallach et al., 2014; Kang et al., 2015; Zhong et al., 2016; Deretic and Levine, 2018), the known instances of plant ectopic cell death seem to involve very different extracellular factors depending on the context (Endo et al., 2001; Tuominen et al., 2004; Overmyer et al., 2005; Wrzaczek et al., 2009, 2015; Escamez et al., 2016). During the formation of the plant vascular xylem tissue, the water-conducting tracheary element (TE) cells rid themselves of their protoplasts to form pipe-like structures by undergoing developmental cell death (Escamez and Tuominen, 2014). Dying TEs release molecules that can be harmful to the neighbouring non-TE cells, and restricting subsequent ectopic non-TE cell death requires active regulation (Endo et al., 2001; Escamez et al., 2016). We had previously found that ectopic non-TE cell death increased upon RNAi-based down-regulation of the TE-expressed Arabidopsis caspase structural homolog METACASPASE9 (MC9) in xylem-differentiating cell suspensions (Escamez et al., 2016).
MC9 down-regulation was also associated with an apparent increase in autophagy specifically in the TE cells (Escamez et al., 2016). Autophagy is an evolutionarily conserved cellular process that targets cellular components for degradation, either in bulk or in a targeted manner, as a means of post-translational regulation or for recycling purposes (Bozhkov, 2018; Marshall and Vierstra, 2018). Strikingly, TE-specific down-regulation of the autophagy protein ATG2 in the MC9-down-regulated cells was found to reduce ectopic non-TE death back to wild-type levels (Escamez et al., 2016). It therefore seems that ectopic non-TE cell death is regulated by the level of autophagy in TEs, downstream of MC9 (Escamez et al., 2016). The mechanism behind this apparent regulation of autophagy by MC9 remains unknown. None of the core autophagy machinery proteins (Marshall and Vierstra, 2018) appears among the identified targets from MC9-mediated proteolysis, and only a single isoform of one autophagy protein (ATG18b) contains putative MC9 target sites (Tsiatsiani et al., 2013). Arabidopsis MC9 and its orthologues in other species have glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an evolutionarily conserved target (Tsiatsiani et al., 2013). GAPDH has been implicated in the regulation of autophagy in several species (Colell et al., 2007; Tristan et al., 2011; Henry et al., 2015), but whether GAPDH could represent the missing link between MC9 and autophagy, upstream of ectopic cell death, has not been investigated.
Ectopic non-TE death in xylem-differentiating cell suspensions is regulated by MC9 and autophagy in TEs, which implies that ectopic non-TE cell death is regulated in a non-cell-autonomous manner. We therefore sought to identify downstream, extracellular factors that mediate the MC9- and autophagy-dependent regulation of ectopic non-TE death. With MC9 being a protease, and autophagy being involved in secretion of proteins and peptides (e.g. during inflammation in animals) (Kimura et al., 2017a,b; Zhang et al., 2017), we hypothesized that extracellular peptides could function to regulate ectopic non-TE death downstream of MC9 and autophagy.
Materials and methods
Plant material and growth conditions
Cell suspensions from the Col-0 wild-type background were previously described (Escamez et al., 2016; Pesquet et al., 2010). MC9-RNAi cell suspensions were generated as previously described (Escamez et al., 2016) by Agrobacterium-mediated transformation of wild-type cell suspensions. Growth conditions and induction of vascular xylem-like differentiation were as previously described (Escamez et al., 2016).
Arabidopsis genotypes were all in the Col-0 background. The T-DNA knockout line mc9-2 (SALK_075814) has been previously published (Bollhöner et al., 2013). The T-DNA lines kratos-1 (SALK_201112) and bia-1 (SALK_069212) were obtained from the Nottingham Arabidopsis Stock Centre (NASC), and homozygous plants were identified that did not display expression of the corresponding genes (Supplementary Fig. S1 at JXB online). For identification of knock-out mutants kratos-1 and bia-1, the RNA was isolated with RNeasy plant kit (Qiagen), cDNA synthesis was performed with QuantiTect reverse transcription kit (Qiagen), and semi-quantitative PCRs were run with Go Taq Green Master Mix (Promega) according to manufacturer’s instructions (Bia forward primer: ATGACTCGAGGAAGTCAAAG; Bia reverse primer: TCACTTATTGTTTCCTTTGCCT; Kratos forward primer: ATGGGGCGTCTCGTTAGTG; Kratos reverse primer: TTAGTGGTGTCCGATTCCG). The transcriptional reporter lines proMC9::GUS and proIRX1::GFP:GUS were previously described (Bollhöner et al., 2013; Escamez et al., 2016). The vascularly expressed autophagasomal fluorescent marker proATG8a::GFP:ATG8a and the fluorescently labelled MC9 under the transcriptional control of the MC9 endogenous promoter proMC9::MC9:mCherry have been previously described (Bollhöner et al., 2013; Furuta et al., 2014).
The overexpressor of the autophagy rate-limiting ATG5 protein (Minina et al., 2018) in TEs proIRX1::ATG5 was generated as follows. The ATG5 full coding sequence was amplified by PCR, using primers including sequences for Gateway BP recombination (forward: GGGGACAAGTTTGTACAAAAAAGCAGCAGGCTTAATGGCG AAGGAAGCGGTCAAG; reverse: GGGGACCACTTTGTACAAGA AAGCTGGGTATCACCTTTGAGGAGCTTTCAC), and recombined using BP clonase into pDONR207 to generate a Gateway-compatible pENTR vector. The IRX1 promoter fragment (proIRX1; 1586 bp upstream of IRX1/AT4G18780) had previously been inserted instead of the 35S promoter fragment in the Gateway-compatible destination vector pK2GW7 (Karimi et al., 2002; Escamez et al., 2016). This proIRX1-containing destination vector was recombined with the ATG5-containing pENTR vector using LR clonase, resulting in the proIRX1::ATG5 binary vector. Agrobacterium tumefaciens (stain GV3101) cells were electroporated with the proIRX1::ATG5 binary vector, and proIRX1::ATG5 Arabidopsis plants were generated by Agrobacterium-mediated transformation. proIRX1::ATG5 proATG8a::GFP:ATG8a was generated by crossing.
For induction of vascular differentiation in cotyledons using the Vascular Cell Induction Culture System Using Arabidopsis Leaves (VISUAL) system, both growth and induction conditions were extensively described previously (Kondo et al., 2016). Each induction of one genotype in one treatment condition included four biological replicates (four seedlings in one and the same well). Plants used for stress-induced cell death assays were grown on soil (peat: vermiculite=1:1) under white luminescent light (220–250 µmol m−2 s−1) with a 12 h photoperiod (temperature 23/18 °C, relative humidity 70/90%) for 5 weeks.
Isolation of extracellular peptides
Wild-type and MC9-RNAi cell suspensions were induced to differentiate (or not, as control) in triplicates, each replicate being in a volume of 200 ml (in 1 litre Erlenmeyer flasks). After 5 d, when the rate of TE differentiation is peaking (Escamez et al., 2016), concomitantly with the rise of ectopic non-TE death in MC9-RNAi (Escamez et al., 2016), the samples were harvested. The cells and extracellular medium of each sample were separated using inert sterile membranes with a 0.45 µm pore size (Millipore Stericup-HV Filter Units). The extracellular medium fractions were further filtered using Jumbosep centrifugation devices with a 3 kDa cut-off membrane (Pall) to separate the peptides (<3 kDa) from higher molecular mass (>3 kDa) protein (fragments). The higher molecular mass extracellular fraction was compared with cell lysates to verify the absence of detectable intracellular contaminations in the extracellular medium (Supplementary Fig. S2A). Peptides in the low molecular mass (<3 kDa) extracellular fraction were further purified by reverse-phase C18 solid phase extraction (SepPak C18, Waters) and the resulting samples were freeze-dried.
Peptide identification and relative quantification
Freeze-dried peptide samples were dissolved in 10 µl of 50% acetonitrile (ACN) and subsequently diluted in 200 µl ultrapure water and acidified with trifluoroacetic acid (TFA) to a 1% (v/v) final concentration. No digestion by trypsin or other enzymes was performed, this to preserve the structure of the endogenously generated peptides. Ten microlitres of each sample was subjected to liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis using a Q Exactive mass spectrometer (Thermo Fisher Scientific) that was operated as previously described (Stes et al., 2014).
Raw mass spectrometry data were searched against TAIR10 representative proteins and contaminant protein sequences by the built-in Andromeda search engine of the MaxQuant software (v1.5.3.30) (Cox and Mann, 2008). The ‘unspecific cleavage’ setting was selected in MaxQuant as no protease was used to prepare the samples. The minimum peptide length was set to eight amino acids, matching-between-runs was enabled with an alignment time of 30 min and matching time window of 30 s. Variable modifications were oxidation of methionine and proline (+15.995 Da) and protein N-terminal acetylation (+42.011 Da). The identified peptides (false discovery rate <0.01) and their respective intensities were extracted from the ‘peptides.txt’ output file. As trimming of N- and C-terminal ends of peptides was frequently observed, we grouped overlapping peptides to a single longest peptide variant (LPV) (Secher et al., 2016). More specifically, if a peptide sequence was contained within another peptide or extended another peptide by up to three amino acids, both peptides were grouped to a single LPV. The intensity of a LPV is the sum of the intensities of its constitutent peptides.
In the next phase, the Perseus (v1.5.4.0) computational platform (Tyanova et al., 2016) was used for quantitative analysis of the peptidomics data. Peptides matching contaminants, decoy proteins, or without an intensity value were omitted, resulting in a list of 689 peptides (Supplementary Dataset S1). For statistical analyses, only peptides with at least intensity values for two biological replicates in at least one condition were included, resulting in 201 unique peptides compared with statistical tests. Peptide intensities were log10 transformed and missing values were imputed for the total matrix from the normal distribution with a downshift of 1.8 and width of 0.3. A two sample Student’s t-test was performed for pairwise comparisons and the interaction between the condition (induced or not) and genotype effects was tested by a two-way ANOVA.
The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the dataset identifier PXD010886.
Preparation of peptide solutions
Crude, unpurified (35%<purity<70%, depending on the peptide) peptide synthesis solutions for all the candidate peptides (Supplementary Table S1) as well as high purity peptide solutions (>95%) for peptides 3, 4, Kratos and Bia were obtained from GeneCust. All the peptides were dissolved in 10 mM phosphate buffer pH 7 to 1 mM master stocks, taking into account peptide purity in the crude extract to actually obtain 1 mM peptide solutions. Whenever a peptide was visibly insoluble at pH 7, the pH of the master solutions was either lowered with formic acid or increased with ammonia until dissolution was observed. From the 1 mM peptide solutions, master aliquots of 1, 10 and 50 µM (1000× solutions) were prepared for experimental use without repeating freezing–thawing cycles.
Microscopy and image analyses
Samples for promoter–reporter assays with β-glucuronidase (GUS) staining were prepared as previously described (Bollhöner et al., 2013). All other cotyledon samples (VISUAL) were fixed and cleared prior to microscopy observation as previously described (Kondo et al., 2016).
All micrographs used for display and quantifications were acquired using an Axioplan II epifluorescence upright microscope equipped with an Axiocam HR color camera. The same imaging settings were conserved for all fluorescence micrographs used for quantifications of TE differentiation and ectopic non-TE death. For instance, images were acquired with a ×4 lens, a SOLA SM II LED light source was used for excitation, a dichroic beam splitter allowed blue light excitation (450–490 nm) and long pass detection of all emission fluorescence from green to red (long pass from 515 nm onward), and an exposure time of 380 ms was used.
Quantification of cotyledon areas covered with GUS staining, with TEs, or with dead non-TEs was performed by manually selecting and measuring the corresponding regions of interest (ROIs) in the open source software ImageJ. Especially, identification of TE and dead non-TE autofluorescence required manual input from the experimenters due to the fact that the numerous variable features of autofluorescence (cell size, cell shape, layering of cells, autofluorescence intensity, patterned secondary walls, etc.) did not allow for fully automatized segmentation pipelines (Supplementary Fig. S3N, O). As a result, autofluorescent dead non-TEs were manually identified and selected using the ‘selection brush tool’ in ImageJ, by several experimenters in parallel and with high number of experiment replication to ensure reproducibility. The selected dead non-TE autofluorescence area was quantified with ImageJ, as well as the total autofluorescence area of each cotyledon, allowing us to deduce the TE autofluorescence area by subtracting the dead non-TE area from the total autofluorescence area.
Confocal laser scanning microscopy analyses were performed as described previously (Escamez et al., 2016), using an inverted Zeiss LSM780 confocal microscope. Imaging for colocalization purposes was performed with a ×40 water immersion lens by simultaneous excitation (due to the rapid movement of autophagosomes) of green fluorescent protein (GFP) and mCherry with 488 nm and 561 nm lasers, respectively, with a MBS 488/561 beam splitter, and emission detection windows of 495–546 nm and 595–656 nm, respectively. Colocalization analyses were performed using ImageJ with the Pearson–Spearman Colocalization (PCS) plugin. For counting of GFP:ATG8a puncta, all compared images within an experiment were acquired with the same settings. To distinguish the puncta from the diffuse GFP fluorescence background, a single threshold for fluorescence intensity in the GFP channel was applied to all the images from a single experiment.
Stress-induced cell death assays
Mechanical induction of cell death was performed similarly to a previously described study (Wrzaczek et al., 2009). Briefly, fully expanded leaves of 5-week-old Col-0 or mc9-2 plants were infiltrated with 50 nM Kratos, Bia, or phosphate buffer as a control. Leaf disks were cut from infiltrated leaves, thereby mechanically inducing cell death. Each leaf disk was placed in 5 ml water, allowing for measurement of ion leakage with a conductivity meter. For mitogen-activated protein kinase (MAPK) assays, infiltrated leaves were snap frozen in liquid nitrogen 0, 5, 15, and 30 min after infiltration with 50 nM Kratos, Bia, or phosphate buffer.
Induction of cell death by oxidative stress using the so-called xanthine/xanthine oxidase (X/XO) system (Jabs et al., 1996) has previously been described (Overmyer et al., 2005). Briefly, fully expanded leaves of 5-day-old Col-0, kratos-1, and bia-1 plants were detached and infiltrated with a buffer containing a superoxide-generating xanthine/xanthine oxidase mixture, or only buffer as a control. After 4 h, the detached leaves were rinsed and cell death from each leaf was quantified by placing the detached leaf in 5 ml water where ion leakage was measured with a conductivity meter, immediately (+0 h) or after another 4 h (+4 h).
Reactive oxygen species burst measurements
Leaf discs were collected using a 4 mm cork borer from 4-week-old Arabidopsis Col-0 plants and floated overnight in sterile distilled water in a 96-well plate under continuous light at room temperature. On the following day, the water was replaced with assay buffer containing 34 mg l−1 Luminol sodium salt (Sigma), 20 mg l−1 horseradish peroxidase (Wako), 100 nM flg22 (GenScript), or synthetic peptides. Luminescence was measured using the GloMax®-Multi+Detection System (Promega). ROS production was expressed in relative luminescence units (RLU). Data are presented as the average of six leaves in a representative experiment and the experiment was repeated three times with similar results.
Immunoblotting
Extracellular medium containing proteins >3 kDa after filtration was concentrated using Amicon centrifugal filter units (10 kDa cut-off, Millipore). The control cells were lysed in urea buffer (6 M urea, 50 mM Tris–HCl, pH 7.5, protease inhibitor cocktail 1× (Sigma-Aldrich)). Equal protein amounts (20–50 µg) separated by SDS-PAGE were transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). The membranes were blocked with 5% milk and probed with anti-HSP101 or anti-GDC-H antibody (1:1000 in 1% milk, TBS-T) (Agrisera). Horseradish peroxidase-conjugated donkey anti-rabbit IgG (GE Healthcare) was used as a secondary antibody and the signal was visualized by ECL Prime luminescence reagents (GE Healthcare).
For MAPK assay the frozen leaves were ground in liquid nitrogen to fine powder and the proteins were extracted by incubation for 30 min at 4 °C in extraction buffer (50 mM HEPES, pH 7.4, 50 mM NaCl, 10 mM EDTA, protease inhibitor cocktail (1×, Sigma-Aldrich), Halt phosphatase inhibitor cocktail (1×, Thermo Fisher Scientific)) with occasional vortexing. The supernatant after centrifugation at 16 000 g for 10 min at 4 °C was used for immunoblotting. One hundred micrograms of total protein was separated on 12% SDS-PAGE and transferred to PVDF membrane. The membrane was blocked with 5% milk in TBS and probed with phospho-p44/42 MAPK antibody (Cell Signaling Technology, no. 4370; 1:2000 in 1% milk, TBS-T). The signal was detected using fluorescent secondary antibody IRDye800CW Goat anti-Rabbit IgG (LI-COR, no. 926-32211) with an Odyssey fluorescence scanner (LI-COR Biosciences). Quantification of band intensity was performed with ImageJ.
GAPDH cleavage analysis
GAPDH purified from rabbit muscle was purchased from Sigma-Aldrich (no. G5262, GAPDH standard for protein electrophoresis and no. G2267, GAPDH for enzymatic purposes). Recombinant Arabidopsis MC9 (rMC9) and mutated inactive rMC9C/A (alanine substitution for the active site cysteine) fused to a His-tag were expressed and purified from Escherichia coli as previously described (Vercammen et al., 2004). GAPDH proteins were dissolved at 2 mg ml−1. A dilution series of rMC9 in Milli-Q purified water was made starting from 1/25 (w/w) ratio. A 40 µl reaction mix consisted of 10 µl 4×MC9 buffer (200 mM MES pH 5.5, 600 mM NaCl, 40% sucrose, 0.4% CHAPS, and 40 mM DTT), 5 µl GAPDH protein, and 5 µl of rMC9 or rMC9C/A. rMC9 was added to the reaction mix first to auto-activate at room temperature. After 10 min, GAPDH protein was added and incubated for 30 min at 30 °C. The reaction was stopped by the addition of 10 µl 5× Laemmli buffer and samples were run on a 4–20% gradient SDS-PAGE gel (Mini-Protean TGX, Bio-Rad), stained with Instant Blue stain (Expedeon) and imaged with a ChemiDoc Imaging System (Bio-Rad).
Results
Quantitative peptidomics reveals extracellular peptides related to vascular xylem differentiation
To identify extracellular peptides in the context of vascular xylem differentiation, we used a cell suspension system in which hormonal induction triggers semi-synchronous vascular xylem differentiation over a period of 10 d (Pesquet et al., 2010; Escamez et al., 2016). Triplicate wild-type and MC9-down-regulated (MC9-RNAi) cell suspensions were induced to differentiate into xylem TEs and non-TEs, and cells were separated from the extracellular medium by filtration after 5 d when ectopic cell death starts increasing in MC9-RNAi lines (Escamez et al., 2016). Control samples were collected from non-induced wild-type and MC9-RNAi cell suspensions. Proteins were isolated from the extracellular medium and peptides were separated from the higher molecular mass fraction by filtration over a 3 kDa cut-off filter (Fig. 1A). Immunoblotting of the higher molecular mass fraction (>3 kDa) showed no visible contamination with intracellular proteins (Supplementary Fig. S2A). The peptide-containing fraction (<3 kDa) was enriched and purified by solid phase extraction (SPE) (Fig. 1A). Subsequently, peptides were identified and quantified by LC-MS/MS using a Q Exactive mass spectrometer resulting in the identification of 1229 different peptides. Overlapping peptides matching to the same protein sequence, so-called ragged peptides, were assembled into the longest peptide variant (LPV; Fig. 1B) (Secher et al., 2016). After LPV assembly, 722 unique peptides were identified, of which 689 were quantified in at least one replicate of one genotype in one condition (Supplementary Dataset S1).
Fig. 1.
Peptidomics approach to identify METACASPASE9-regulated extracellular peptides. (A) Schematic representation of the bioactive peptide identification workflow. The extracellular media of differentiating cell suspensions were sampled 5 d after induction (n=3 biological replicates), half-way through the differentiation when MC9 expression peaks and when ectopic cell death starts increasing in MC9-RNAi lines (Escamez et al., 2016). (B) Visual explanation for the longest peptide variant (LPV) approach. (C) Venn diagram showing the number of unique peptides per genotype and per condition (induced or non-induced control).
The identified peptides showed a bias in amino acid composition, favouring Gly and Pro (Supplementary Fig. S2B). At the P1 position (position of the precursor protein sequence where cleavage releases the peptide), Gly or Lys was over-represented (Supplementary Fig. S2B). The fact that MC9 cleaves proteins after Arg or Lys (Tsiatsiani et al., 2013) suggests that a subset of the peptides could result from MC9-mediated cleavage.
Induction of xylem differentiation doubled the peptidome repertoire in both wild-type and MC9-RNAi genotypes (Fig. 1C), likely reflecting an increase in proteolytic processes. Under control conditions the extracellular peptidomes of wild-type and MC9-RNAi largely overlapped (185 peptides, ~80% overlap; Fig. 1C), while they diverged more (254 peptides, ~60% overlap; Fig. 1C) after induction of xylem differentiation. Hence, xylem differentiation increased peptide diversity in a genotype-dependent manner. We reasoned that candidate regulator peptides for ectopic non-TE cell death should fulfil three criteria: (i) such peptides should accumulate during in vitro differentiation, (ii) they should be differentially abundant between wild-type (low ectopic death) and MC9-RNAi cells (elevated ectopic death), and (iii) the corresponding genes should be expressed during vascular differentiation. Based on these criteria, we prioritized a set of 15 candidate peptides for further characterization of their bioactivity (Supplementary Table S1).
Kratos restricts runaway ectopic cell death during vascular differentiation
To test the effect of the candidate peptides on ectopic non-TE death, we used the VISUAL system (Kondo et al., 2016) in which differentiation of vascular cells can be induced in the cotyledons (embryonic leaves) of Arabidopsis. This system enables differentiation of TEs within only 4 d (Kondo et al., 2016), as confirmed by the expression patterns of TE marker genes (Bollhöner et al., 2013; Escamez et al., 2016), and by autofluorescence of lignin in the cell walls of TEs (Supplementary Fig. S3A–G). VISUAL also allows monitoring of non-TE cell death. Unlike TEs that become devoid of any visible content because they undergo complete protoplast autolysis (Escamez and Tuominen, 2017), ectopically dying non-TEs retain their protoplasts which become autofluorescent (Supplementary Fig. S3H–J). Autofluorescence of non-TE protoplasts was also observed following mechanical wounding of the cotyledons (Supplementary Fig. S3K–M), suggesting that protoplast autofluorescence could be used as a general hallmark of ectopic cell death in the VISUAL system.
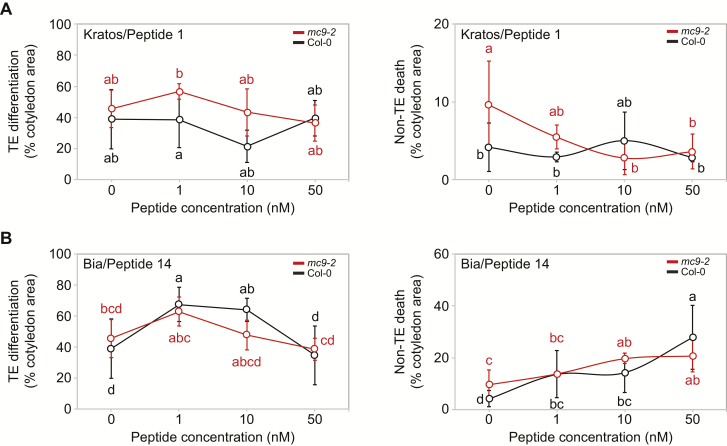
In a first screen, crude synthetic peptide solutions (i.e. without purifying the synthesized peptides) of the 15 candidate peptides were applied to the cotyledons of wild-type (Col-0) and MC9 knockout mutant mc9-2 (Bollhöner et al., 2013) seedlings 24 h after induction of vascular differentiation (Fig. 2A, B; Supplementary Fig. S4). At this stage the newly transdifferentiated pro-cambial cells start differentiating into other vascular cell types such as TEs (Kondo et al., 2016). In agreement with previous results in MC9-RNAi cell suspensions (Escamez et al., 2016), mc9-2 mutants showed significantly more ectopic non-TE death than wild-type seedlings (Fig. 2A, B; Supplementary Fig. S4). Application of three peptides (Peptide 1, Peptides 3 and 4; Supplementary Table S1) led to a reduction of ectopic non-TE death in mc9-2 to levels comparable to wild-type, without any dose-dependent effect on TE differentiation (Fig. 2A; Supplementary Fig. S4). Peptide 1, but not Peptide 3 or Peptide 4, reduced ectopic non-TE death in a dose-dependent manner (Fig. 2A). An opposite, dose-dependent increase in ectopic non-TE death was observed in both wild-type and mc9-2 genotypes upon treatment with Peptide 14 (Fig. 2B).
Fig. 2.
Unpurified peptides Kratos and Bia affect non-TE death in a dose-dependent manner. (A) TE differentiation (left) and non-TE death (right) 96 h after induction with the VISUAL method of Col-0 wild-type and mc9-2 METACASPASE9 knockout mutant treated with increasing concentrations of Kratos/Peptide 1 (or only phosphate buffer as a control). Error bars represent standard deviation (n=3 biological replicates). Data points that do not share any letter are significantly different according to post-ANOVA Fisher’s test (P<0.05). (B) TE differentiation (left) and non-TE death (right) 96 h after induction with the VISUAL method of Col-0 wild-type and mc9-2 METACASPASE9 knockout mutant treated with increasing concentrations of Bia/Peptide 14 (or only phosphate buffer as a control). Error bars represent standard deviation (n=3 biological replicates). Data points that do not share any letter are significantly different according to post-ANOVA Fisher’s test (P<0.05).
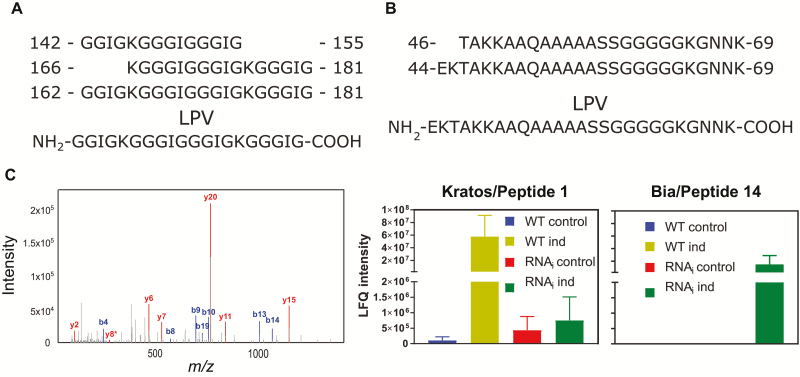
In addition to its dose-dependent effect, the candidate Peptide 1 accumulated in the medium of wild-type cells, but not upon MC9 down-regulation (Fig. 3A, C; Supplementary Dataset S1). Peptide 1 is part of a precursor protein of unknown function that contains a repeated motif of GGG(I/V)GGG(I/F)GK as well as six repeats of Peptide 1 (Supplementary Fig. S5A), suggesting a putative role as a precursor protein for bioactive peptides. The other peptide with a dose response, the candidate Peptide 14, accumulated only in the medium of differentiating MC9-RNAi cells (Fig. 3B, C). The precursor protein of Peptide 14 is part of an uncharacterized protein family founded by the highly conserved small EDRK-rich factor (SERF, previously known as H4F5) (Lefebvre et al., 1995; Roy et al., 1995; Kelter et al., 2000) proteins in metazoans (Supplementary Fig. S5B, C). We named Peptide 1 (and its corresponding gene/protein AT3G23450) ‘Kratos’, and Peptide 14 (AT3G24100) ‘Bia’, in reference to the homonymous children of the Styx river separating the worlds of the living and of the dead in the ancient Greek mythology.
Fig. 3.
METACASPASE9-dependent accumulation of the peptides Kratos and Bia. (A) The assembly of the longest peptide variant (LPV) from the extracellular peptides matching the glycine-rich peptide Kratos/Peptide1 derived from the unknown protein AT3G23450 (Supplementary Table S1). (B) The assembly of the LPV from the extracellular peptides matching the small EDRK-rich factor (SERF) peptide Bia/Peptide 14 derived from the uncharacterized protein AT3G24100 (Supplementary Table S1). (C) Illustration of mass spectrometry identification of the longest identified peptide in (A) and charts displaying quantifications of the average label free quantification (LFQ) intensity for the LPV Kratos/Peptide 1 and Bia/Peptide 14 in the different genotypes and conditions. Error bars represent standard error of the mean (n=3 biological replicates).
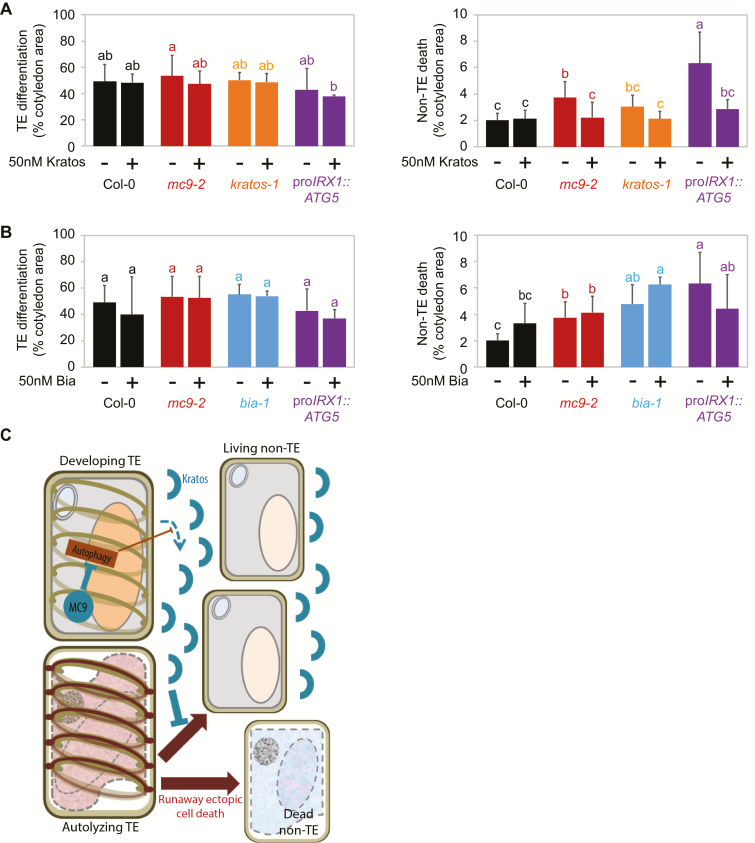
To confirm the cell-survival effect of Kratos, Peptide 3, and Peptide 4, as well as the death-promoting effect of Bia, we repeated the treatments of differentiating cells in the VISUAL system with purified synthetic peptides (purity >95%). Treatment with purified Kratos confirmed its ability to decrease ectopic non-TE death in mc9-2 without affecting TE differentiation (Fig. 4A). Furthermore, a knockout T-DNA line for the Kratos-encoding gene (AT3G23450; kratos-1)—devoid of visible xylem differentiation defect (Supplementary Fig. S6)—showed a trend towards higher ectopic non-TE death than wild-type (Fig. 4A), which could be complemented by treatment with purified Kratos (Fig. 4A). These results, together with the reduced abundance of extracellular Kratos in differentiating MC9-RNAi cell suspensions (Fig. 3C) and the increased ectopic non-TE death upon loss of MC9 function (Figs 2, 4), indicate that Kratos functions in restricting MC9-dependent ectopic non-TE death during xylem differentiation.
Fig. 4.
Ectopic non-TE death is restricted by the Kratos peptide during induced vascular differentiation in cotyledons. (A) TE differentiation (left) and non-TE death (right) 96 h after induction with the VISUAL method of Col-0, mc9-2, kratos-1 knockout mutant and proIRX1::ATG5 TE autophagy inducer line with or without 50 nM Kratos. Error bars represent standard deviation (n=9 replicate experiments for Col-0 and mc9-2, 5 for Col-0+Kratos, mc9-2+Kratos, kratos-1 and kratos-1+Kratos, 4 for proIRX1::ATG5, and 3 for proIRX1::ATG5+Kratos, each with three biological replicates). Data points that do not share any letter are significantly different according to post-ANOVA Fisher’s test (P<0.05). (B) TE differentiation (left) and non-TE death (right) 96 h after induction with the VISUAL method of Col-0, mc9-2, bia-1 knockout mutant and proIRX1::ATG5 TE autophagy inducer line with or without 50 nM Bia. Error bars represent standard deviation (n=9 replicate experiments for Col-0 and mc9-2, 8 for Col-0+Bia and mc9-2+Bia, 4 for proIRX1::ATG5, and 3 for bia-1, bia-1+Bia and proIRX1::ATG5+Bia, each with three biological replicates). Data points that do not share any letter are significantly different according to post-ANOVA Fisher’s test (P<0.05). (C) Hypothetical model for the modulation of non-TE ectopic death by MC9 and autophagy in TEs through regulation of extracellular accumulation of the Kratos peptide.
Peptides 3 and 4 were disqualified as potential regulators of ectopic non-TE death based on the inability of the corresponding purified peptides to significantly decrease ectopic non-TE death in mc9-2 (Supplementary Fig. S7), and because T-DNA lines for the corresponding genes did not display consistent changes in ectopic death (Supplementary Fig. S7). The potential role of Bia in promoting ectopic non-TE death during vascular differentiation remained unclear because treatment with the corresponding purified peptides produced inconsistent effects depending on the treated genotype (Fig. 4B). Furthermore, even without treatment ectopic non-TE death tended to increase, rather than decrease, in a T-DNA knockout line for the Bia-encoding gene (AT3G24100; bia-1; Fig. 4B).
Kratos functions downstream of autophagy in TEs
Autophagy has been suggested to increase in the TEs of MC9-down-regulated cell suspensions, which also display elevated ectopic non-TE death (Escamez et al., 2016). Cell-type-specific down-regulation of the autophagy gene ATG2 to decrease autophagy in TEs was found sufficient to restrict ectopic non-TE death in MC9-down-regulated cell suspensions (Escamez et al., 2016). Consistent with the modulation of autophagy by MC9 in TEs, we found a partial co-localization between MC9 and the vascularly expressed autophagosome marker ATG8a (Furuta et al., 2014) in TEs in roots of Arabidopsis seedlings (Supplementary Fig. S8A). Visualization of autophagosomes with the same marker showed increased autophagy in the TEs of mc9-2 compared with the wild-type (Supplementary Fig. S8B). Furthermore, the extracellular medium of xylem differentiating cell suspensions contained six unique peptides matching three subunits of the GAPDH protein (Supplementary Dataset S1), a known regulator of autophagy (Colell et al., 2007; Tristan et al., 2011; Henry et al., 2015), as well as an evolutionarily conserved target of MC9 and its orthologues (Tsiatsiani et al., 2013). Five of the six peptides from GAPDH precursor proteins contain potential MC9 cleavage sites (Supplementary Fig. S9A). Mammalian GAPDH could be cleaved in vitro by recombinant Arabidopsis MC9 in a dose-dependent manner, while no cleavage was observed in the absence of MC9 or in the presence of catalytically inactive MC9 (Supplementary Fig. S9B). Sequence alignment between the assayed mammalian GAPDH and the Arabidopsis GAPDH proteins showed a high degree of conservation, including conservation of three potential MC9 cleavage sites (Supplementary Fig. S9C) between the detected Arabidopsis GAPDH-derived peptides (Supplementary Fig. S9A) and the assayed mammalian protein (Supplementary Fig. S9C). It is therefore reasonable to hypothesize that MC9 regulates autophagy in TEs by cleaving GAPDH, which would also lead to the release of GAPDH peptides during TE autolysis, thereby explaining that we detected extracellular GAPDH-derived peptides.
Given that TE autophagy modulates ectopic non-TE death downstream of MC9, it is possible that modulation of autophagy in TEs could function upstream of the Kratos-mediated restriction of ectopic non-TE death. To test this hypothesis, we increased the level of autophagy in TEs by overexpressing the autophagy rate-limiting protein ATG5 (Minina et al., 2018) under the transcriptional control of the IRX1 promoter (proIRX1::ATG5), which is active in TEs and not in non-TEs (Escamez et al., 2016) (Supplementary Fig. S3A–G). Overexpression of ATG5 in TEs increased autophagy in these cells (Supplementary Fig. S8C) without significant effect on TE differentiation compared with wild-type (Fig. 4A, B). Instead, consistent with a link between TE autophagy and ectopic non-TE death, ATG5 overexpression in TEs resulted in a significant increase in ectopic non-TE death (Fig. 4A, B). Ectopic non-TE death in proIRX1::ATG5 cotyledons was decreased to wild-type levels by treatment with Kratos peptide (Fig. 4A). Given that extracellular abundance of Kratos was reduced in cell suspensions where TEs displayed MC9 down-regulation and hence increased autophagy (Fig. 3C), the restriction of ectopic non-TE death in proIRX1::ATG5 by Kratos (Fig. 4A) suggests that this peptide functions downstream of TE autophagy (Fig. 4C). Unexpectedly, ectopic non-TE death in proIRX1::ATG5 was decreased upon treatment with Bia (Fig. 4B), suggesting that the relation between Bia and TE autophagy may be complex.
Kratos and Bia modulate ectopic cell death induced by stress
The ability of Kratos (Figs 2A, 4A) to modulate ectopic cell death during induction of vascular differentiation, and the possible death-promoting effect of Bia (Fig. 2B), prompted us to test whether these peptides could modulate ectopic cell death in other contexts than vascular differentiation.
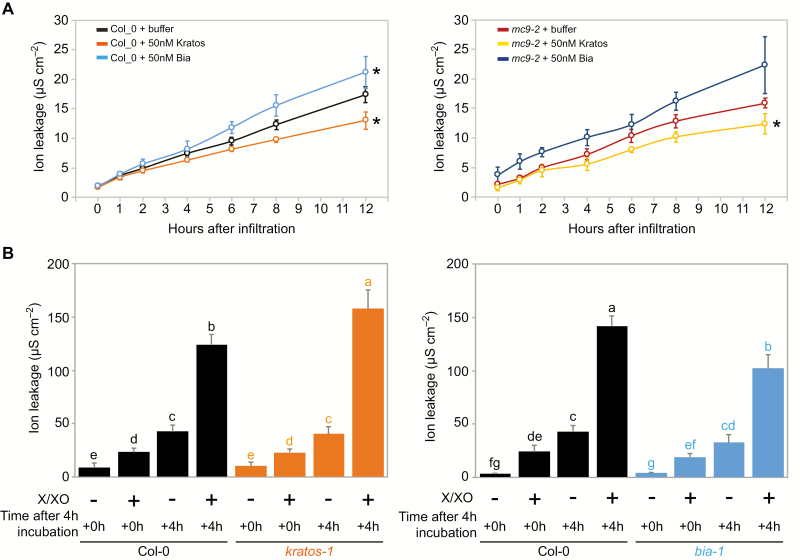
Mechanical stress was imposed to induce wounding and subsequent ectopic cell death by excision of leaf disks from wild-type and mc9-2 leaves and infiltration with buffer alone or with peptide. Indeed, following local mechanical stress, cell death has been shown to spread from the site of application to other cells (Greenberg and Ausubel, 1993; Wrzaczek et al., 2015). Here, cell death was estimated over time by measuring electrolyte leakage (Fig. 5A). Electrolyte leakage dynamics were comparable between wild-type and mc9-2 (Fig. 5A), as expected from the fact that MC9 is expressed in cells undergoing developmental, rather than accidental, cell death (Olvera-Carrillo et al., 2015). Cell death increased in both genotypes upon treatment with the Bia peptide (Fig. 5A), consistent with an ectopic cell death-promoting role. Treatment with the Kratos peptide decreased cell death following mechanical stress regardless of the genotype (Fig. 5A), consistent with the ability of Kratos to restrict ectopic cell death.
Fig. 5.
Peptides Kratos and Bia have the ability to modulate stress-induced cell death. (A) Mechanically induced cell death (measured by ion leakage) in leaf disks from Col-0 wild-type or mc9-2 leaves infiltrated with Kratos or Bia peptides, or with buffer as a control. Data points indicate average values while error bars represent standard deviation (n=4 biological replicates). Asterisks indicate significantly different (P<0.05) ion leakage dynamics compared with buffer control. (B) Cell death induced by 4 h treatment with superoxide-generating xanthine/xanthine oxidase (X/XO) and quantified by measuring ion leakage in detached leaves from Col-0, kratos-1, and bia-1 0 h (+0 h) or 4 h (+4 h) after the end of the 4 h treatment. Bars indicate average values while error bars represent standard deviation (n=4 biological replicates). Data points that do not share any letter are significantly different according to post-ANOVA Fisher’s test (P<0.05). The experiments have been repeated twice with similar results.
Additionally, cell death was induced by extracellular reactive oxygen species (ROS) production in leaves of wild-type as well as in the null mutants kratos-1 and bia-1, and cell death was measured by electrolyte leakage after 4 h (Fig. 5B). Cell death in response to extracellular ROS production was significantly higher in kratos-1 plants compared with wild-type (Fig. 5B), consistent with an anti-ectopic cell death role for Kratos. The ability of Bia to enhance cell death was also supported by the fact that bia-1 plants displayed less ectopic cell death than wild-type plants (Fig. 5B).
Kratos and Bia regulated cell death in ways that are reminiscent of responses to microbe-associated molecular patters (MAMPs) and/or damage-associated molecular patterns (DAMPs) (Bigeard et al., 2015). MAMPs and DAMPs trigger signalling cascades whose hallmarks include the production of extracellular ROS and activation of mitogen-activated protein kinases (MAPK or MPK), especially MPK3 and MPK6 (Bigeard et al., 2015). We investigated whether Kratos and Bia could modulate ectopic cell death through similar signalling networks. Unlike the MAMP flg22, an epitope of the bacterial flagella which triggers an oxidative burst, neither Kratos nor Bia application triggered a ROS burst (Supplementary Fig. S10). On the other hand, MPK3 and MPK6 activation, which typically peaks within 10–15 min after signal perception (Bigeard et al., 2015), was higher 15 min after infiltration of Arabidopsis leaves with Kratos or Bia compared with controls (Supplementary Fig. S10). Together these results suggest that some downstream responses to Kratos and Bia could be shared with MAMPs or DAMPs while others seem to be distinct.
Our collective results reveal that Kratos has the ability to restrict ectopic cell death both in a normal developmental context and as a consequence of stress, while Bia conclusively promotes ectopic cell death only in the tested stress conditions. At least during xylem differentiation and wounding, cell death can spread in a regulated manner, which reveals that the original trigger itself does not necessarily cause all of the observed death events. Instead, the cell death that ectopically spreads likely represents a specific type of cell death with its own genetic regulation (Wrzaczek et al., 2009; Wrzaczek et al., 2015) (Figs 4A, 5) that relies on extracellular peptides (Figs 3–5), one of which apparently functions downstream of autophagy in TEs (Fig. 4A).
Discussion
By combining peptidomics with the use of a controlled plant vascular differentiation system, we identified 1229 extracellular peptides, corresponding to 433 proteins, in the context of plant vascular development (Supplementary Dataset S1). These numbers, as often with peptidomics, differ from current high mass-accuracy shotgun proteomics. Nevertheless, our analysis performed at least as well as other label-free peptidomics (Nanni et al., 2009; Ling et al., 2010; Chen et al., 2014) studies when considering the caveats associated with endogenous peptidomics (Dallas et al., 2015). One frequently reported challenge in peptidomics is the occurrence of progressively shortened N- or C-terminal peptide ends. Assuming that they belong to the same (bioactive) precursor, we aggregated such ragged peptides to a single longest peptide variant (LPV; Fig. 1B), thereby facilitating data interpretation and enhancing label-free quantification as the intensity of the LPVs is the sum of the intensities of all their nested peptides. A previous proteomics study identified 149 proteins with an orthologue in Arabidopsis in the xylem sap of Brassica oleracea (Ligat et al., 2011) but did not focus on the peptide fraction. While the different methods and data processing pipelines between both studies render direct comparison of the detected peptides challenging (Ligat et al., 2011), 27 proteins identified in the xylem sap of Brassica oleracea matched precursor proteins of the peptides identified in our dataset (Supplementary Dataset S1). This suggests that our dataset is relevant for the developmental context of vascular xylem differentiation.
Differentiating TEs undergo developmentally regulated cell death and protoplast autolysis (Escamez and Tuominen, 2014), during which they release proteases, and likely other molecules, which can either directly injure and kill neighbouring cells (Endo et al., 2001) or possibly activate signalling towards ectopic cell death (Endo et al., 2001; Escamez et al., 2016). To prevent ectopic non-TE death, it was shown that at least one protease inhibitor must be released in the extracellular medium of xylem-differentiating cell suspensions in Zinnia elegans (Endo et al., 2001). We report here the extracellular peptide Kratos as another factor that is required to restrict ectopic non-TE death in Arabidopsis. Kratos was abundant in wild-type xylem-differentiating cell suspensions (Fig. 3C), and it rescued the ectopic cell death during vascular differentiation in the mc9-2 mutant (Figs 2A, 4A). Another peptide, Bia, was only detected in the extracellular medium of differentiating, ectopic cell death-prone MC9-RNAi cell suspensions (Fig. 3C) and displayed a tendency to promote ectopic cell death following stress (Fig. 5A, B). Our data suggests that ectopic non-TE death is modulated by a balance between pro-death and pro-survival factors. This balance is altered when MC9 is down-regulated (Fig. 3C; Supplementary Dataset S1), resulting in enhanced ectopic death (Escamez et al., 2016) (Figs 4, 5). We identified the extracellular peptide Kratos as one of these factors regulating runaway ectopic cell death, and possibly also Bia. Neither of the precursor proteins for Bia or Kratos had been found among MC9 targets in Arabidopsis seedlings and their cleavage sites do not display the MC9-specific lysine or arginine amino acid at the P1 position. Hence, Kratos and Bia are likely indirect targets of MC9, possibly through MC9-dependent modulation of TE autophagy (Supplementary Fig. S8A, B) (Escamez et al., 2016), as seems to be the case for Kratos (Figs 3C; 4A).
An important function of MC9 in TEs during vascular development is the regulation of autophagy (Escamez et al., 2016) (Supplementary Fig. S8B), which in turn regulates non-TE ectopic cell death (Escamez et al., 2016) (Fig. 4A–C). More precisely, our results indicate that MC9 tunes down the level of autophagy in TEs, while TE autophagy negatively regulates the extracellular accumulation of the ectopic cell death-restricting Kratos peptide (Fig. 4C). Frequent identification of peptides derived from GAPDH (Supplementary Dataset S1), which is known to regulate autophagy (Colell et al., 2007; Tristan et al., 2011; Henry et al., 2015) and can be cleaved by MC9 (Tsiatsiani et al., 2013) (Supplementary Fig. S9B), allows us to hypothesize that GAPDH cleavage by MC9 could mediate the modulation of autophagy upstream of ectopic cell death.
Autophagy is known as a cellular process in which double-membrane-enclosed autophagosome vesicles transport cellular components to vacuoles or lysosomes for degradation and recycling (Marshall and Vierstra, 2018), which seems contradictory to a role in regulating accumulation of extracellular peptides. Yet, studies in animals have found a role for autophagy in secreting proteins or peptides (Bhattacharya et al., 2014; Kimura et al., 2017b), which has also been hypothesized in plants (Pečenková et al., 2017). Hence, TE autophagy may directly regulate extracellular peptide accumulation similar to the secretion of the mammalian pro-inflammatory interleukin-1β peptide by autophagy (Dupont et al., 2011). TE autophagy may also play a role in degrading an upstream regulator of extracellular peptide accumulation, as observed for mammalian senescence-associated secretory phenotype (Kang et al., 2015). A third possibility is that different levels of autophagy in TEs could influence the intracellular content released by TEs upon loss of plasma membrane integrity during developmental cell death and autolysis.
Intriguingly, while identified in a specific developmental context, both Kratos and Bia possess the ability to modulate ectopic cell death that results from stress. Stress-induced cell death and developmental cell death are known to display distinct transcriptional and anatomical hallmarks (Overmyer et al., 2005; Torres et al., 2005; van Doorn, 2011; Fendrych et al., 2014; Olvera-Carrillo et al., 2015; Escamez and Tuominen, 2017), indicating that they represent different types of cell death. It is therefore surprising to find that Kratos and Bia can modulate ectopic cell death occurrences that follow different triggers. A possible explanation is that, after stress-induced or developmental cell death, the resulting ectopic cell death represents one specific type of cell death with its own anatomical features and molecular machinery, different from the cell death that is induced directly in response to the initial stimulus. This seems to be the case at least for the ectopic non-TE death that is anatomically distinct from the cell death of the TEs (Escamez et al., 2016) (Supplementary Fig. S3H–M), and where only the former and not the latter is affected by Kratos (Escamez et al., 2016) (Figs 2A, 4A). The spreading, runaway cell death in plants that follows stress-induced or developmental cell death may therefore be analogous to an inflammation-like response caused by the initial, ‘primary’ cell death. Given our finding that Kratos can restrict runaway ectopic cell death in developmental and stress-related contexts, we hypothesize the existence of a plant inflammation-like response involving Kratos as a canonical effector.
Supplementary data
Dataset S1. Peptide identification from extracellular medium of cell suspensions differentiating (or not as a control) into a mixture of xylem tracheary elements (TE) and non-TE cells.
Fig. S1. Identification of knockout T-DNA lines for Kratos and Bia.
Fig. S2. Quantitative peptidomics on extracellular medium of xylogenic Arabidopsis cell suspensions.
Fig. S3. Monitoring TE differentiation and non-TE death with the Vascular Cell Induction Culture System Using Arabidopsis Leaves (VISUAL).
Fig. S4. Effect of candidate peptides from crude extracts on TE dierentiation and non-TE death.
Fig. S5. Sequences of the precursor proteins from which the peptides Kratos and Bia are generated.
Fig. S6. Normal vascular development in the cotyledons of the mc9-2, bia-1, and kratos-1 mutants.
Fig. S7. Peptides 3 and 4 do not affect non-TE death.
Fig. S8. METACASPASE9 and autophagy are linked in TEs.
Fig. S9. GAPDH peptides are detected in extracellular peptidomics of differentiating cells, possible from cleavage of GAPDH by MC9.
Fig. S10. Kratos and Bia do not trigger oxidative burst.
Table S1. List of the unique peptides selected for analyses during vascular differentiation.
Author contributions
SE, SS, JPV, MW, and HT conceived the study and obtained or generated the plant material and peptides. SE generated material from cell suspensions for peptide identification. SS, JPV, FVB, and KG performed peptide isolation, enrichment, identification, and data interpretation. SE performed all experiments of xylem differentiation. JPV, HJ, SK, and MW performed stress-induced cell death assays, MPK activity assays, and ROS measurements. SE performed confocal laser scanning microscopy analyses. SE, SS, MW, KG, and HT wrote the manuscript with help from all the other authors.
Acknowledgements
The authors thank Mikko Luomaranta, Maria Baldauf, and Inkeri Soppa for technical assistance. This study was financially supported by European Union 7th Framework Program (PRIME-XS project, grant no. 262067). SS is financially supported by the Flanders Research Foundation (12M3418N). JPV, HJ, SK, and MW are members of the Centre of Excellence in the Molecular Biology of Primary Producers (2014-2019) funded by the Academy of Finland (grant nos 271832 and 307335). MW acknowledges funding from the Academy of Finland (grant nos 275632, 283139, and 312498) and the University of Helsinki (three-year fund allocation). HT is financially supported by the Swedish Research Council VR (grant no. 621-2013-4949) and the Swedish Governmental Agency for Innovation Systems VINNOVA (UPSC Centre for Forest Biotechnology grant no. 2016-00504).
References
- Bhattacharya A, Prakash YS, Eissa NT. 2014. Secretory function of autophagy in innate immune cells. Cellular Microbiology 16, 1637–1645. [DOI] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H. 2015. Signaling mechanisms in pattern-triggered immunity (PTI). Molecular Plant 8, 521–539. [DOI] [PubMed] [Google Scholar]
- Bollhöner B, Zhang B, Stael S, Denancé N, Overmyer K, Goffner D, Van Breusegem F, Tuominen H. 2013. Post mortem function of AtMC9 in xylem vessel elements. New Phytologist 200, 498–510. [DOI] [PubMed] [Google Scholar]
- Bozhkov PV. 2018. Plant autophagy: mechanisms and functions. Oxford: Oxford University Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Pike HM, Olszak B, Skov S, Odum N, Jørgensen LB, Brown RE, Mundy J. 2002. Knockout of Arabidopsis ACCELERATED-CELL-DEATH11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes & Development 16, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Lee CY, Cheng KT, Chang WH, Huang RN, Nam HG, Chen YR. 2014. Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. The Plant Cell 26, 4135–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke PG, Clarke S. 1996. Nineteenth century research on naturally occurring cell death and related phenomena. Anatomy and Embryology 193, 81–99. [DOI] [PubMed] [Google Scholar]
- Colell A, Ricci JE, Tait S, et al. 2007. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell 129, 983–997. [DOI] [PubMed] [Google Scholar]
- Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, Epple P. 2010. Arabidopsis type I metacaspases control cell death. Science 330, 1393–1397. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Dallas DC, Guerrero A, Parker EA, Robinson RC, Gan J, German JB, Barile D, Lebrilla CB. 2015. Current peptidomics: applications, purification, identification, quantification, and functional analysis. Proteomics 15, 1026–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneva A, Gao Z, Van Durme M, Nowack MK. 2016. Functions and regulation of programmed cell death in plant development. Annual Review of Cell and Developmental Biology 32, 441–468. [DOI] [PubMed] [Google Scholar]
- Deretic V, Levine B. 2018. Autophagy balances inflammation in innate immunity. Autophagy 14, 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. 2011. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. The EMBO Journal 30, 4701–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S, Demura T, Fukuda H. 2001. Inhibition of proteasome activity by the TED4 protein in extracellular space: a novel mechanism for protection of living cells from injury caused by dying cells. Plant & Cell Physiology 42, 9–19. [DOI] [PubMed] [Google Scholar]
- Escamez S, André D, Zhang B, Bollhöner B, Pesquet E, Tuominen H. 2016. METACASPASE9 modulates autophagy to confine cell death to the target cells during Arabidopsis vascular xylem differentiation. Biology Open 5, 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamez S, Tuominen H. 2014. Programmes of cell death and autolysis in tracheary elements: when a suicidal cell arranges its own corpse removal. Journal of Experimental Botany 65, 1313–1321. [DOI] [PubMed] [Google Scholar]
- Escamez S, Tuominen H. 2017. Contribution of cellular autolysis to tissular functions during plant development. Current Opinion in Plant Biology 35, 124–130. [DOI] [PubMed] [Google Scholar]
- Fendrych M, Van Hautegem T, Van Durme M, et al. 2014. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Current Biology 24, 931–940. [DOI] [PubMed] [Google Scholar]
- Furuta KM, Yadav SR, Lehesranta S, et al. 2014. Plant development. Arabidopsis NAC45/86 direct sieve element morphogenesis culminating in enucleation. Science 345, 933–937. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Bravo-San Pedro JM, Vitale I, et al. 2015. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death and Differentiation 22, 58–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JT, Ausubel FM. 1993. Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. The Plant Journal 4, 327–341. [DOI] [PubMed] [Google Scholar]
- Henry E, Fung N, Liu J, Drakakaki G, Coaker G. 2015. Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genetics 11, e1005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysmans M, Lema A S, Coll NS, Nowack MK. 2017. Dying two deaths – programmed cell death regulation in development and disease. Current Opinion in Plant Biology 35, 37–44. [DOI] [PubMed] [Google Scholar]
- Inohara N, Nuñez G. 2003. NODs: intracellular proteins involved in inflammation and apoptosis. Nature Reviews. Immunology 3, 371–382. [DOI] [PubMed] [Google Scholar]
- Jabs T, Dietrich RA, Dangl JL. 1996. Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273, 1853–1856. [DOI] [PubMed] [Google Scholar]
- Kang C, Xu Q, Martin TD, Li MZ, Demaria M, Aron L, Lu T, Yankner BA, Campisi J, Elledge SJ. 2015. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 349, aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kelter AR, Herchenbach J, Wirth B. 2000. The transcription factor-like nuclear regulator (TFNR) contains a novel 55-amino-acid motif repeated nine times and maps closely to SMN1. Genomics 70, 315–326. [DOI] [PubMed] [Google Scholar]
- Kimura T, Jia J, Claude-Taupin A, et al. 2017. a Cellular and molecular mechanism for secretory autophagy. Autophagy 13, 1084–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Jia J, Kumar S, et al. 2017. b Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. The EMBO Journal 36, 42–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Nurani AM, Saito C, Ichihashi Y, Saito M, Yamazaki K, Mitsuda N, Ohme-Takagi M, Fukuda H. 2016. Vascular cell induction culture system using Arabidopsis leaves (VISUAL) reveals the sequential differentiation of sieve element-like cells. The Plant Cell 28, 1250–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80, 155–165. [DOI] [PubMed] [Google Scholar]
- Ligat L, Lauber E, Albenne C, San Clemente H, Valot B, Zivy M, Pont-Lezica R, Arlat M, Jamet E. 2011. Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 11, 1798–1813. [DOI] [PubMed] [Google Scholar]
- Ling XB, Lau K, Deshpande C, et al. 2010. Urine peptidomic and targeted plasma protein analyses in the diagnosis and monitoring of systemic juvenile idiopathic arthritis. Clinical Proteomics 6, 175–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshin RA, Williams CM. 1964. Programmed cell death—II. Endocrine potentiation of the breakdown of the intersegmental muscles of silkmoths. Journal of Insect Physiology 10, 643–649. [Google Scholar]
- Lockshin RA, Zakeri Z. 2001. Programmed cell death and apoptosis: origins of the theory. Nature Reviews. Molecular Cell Biology 2, 545–550. [DOI] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT. 2001. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proceedings of the National Academy of Sciences, USA 98, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RS, Vierstra RD. 2018. Autophagy: the master of bulk and selective recycling. Annual Review of Plant Biology 69, 173–208. [DOI] [PubMed] [Google Scholar]
- Minina EA, Moschou PN, Vetukuri RR, et al. 2018. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. Journal of Experimental Botany 69, 1415–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanni P, Levander F, Roda G, Caponi A, James P, Roda A. 2009. A label-free nano-liquid chromatography–mass spectrometry approach for quantitative serum peptidomics in Crohn’s disease patients. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences 877, 3127–3136. [DOI] [PubMed] [Google Scholar]
- Olvera-Carrillo Y, Van Bel M, Van Hautegem T, et al. 2015. A conserved core of programmed cell death indicator genes discriminates developmentally and environmentally induced programmed cell death in plants. Plant Physiology 169, 2684–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K, Brosché M, Pellinen R, Kuittinen T, Tuominen H, Ahlfors R, Keinänen M, Saarma M, Scheel D, Kangasjärvi J. 2005. Ozone-induced programmed cell death in the Arabidopsis radical-induced cell death1 mutant. Plant Physiology 137, 1092–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pečenková T, Marković V, Sabol P, Kulich I, Žárský V. 2017. Exocyst and autophagy-related membrane trafficking in plants. Journal of Experimental Botany 69, 47–57. [DOI] [PubMed] [Google Scholar]
- Pesquet E, Korolev AV, Calder G, Lloyd CW. 2010. The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Current Biology 20, 744–749. [DOI] [PubMed] [Google Scholar]
- Reynolds AS. 2014. The deaths of a cell: how language and metaphor influence the science of cell death. Studies in History and Philosophy of Biological and Biomedical Sciences 48 Part B, 175–184. [DOI] [PubMed] [Google Scholar]
- Roy N, Mahadevan MS, McLean M, Shutler G, Yaraghi Z, Farahani R, Baird S, Besner-Johnston A, Lefebvre C, Kang X. 1995. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell 80, 167–178. [DOI] [PubMed] [Google Scholar]
- Secher A, Kelstrup CD, Conde-Frieboes KW, Pyke C, Raun K, Wulff BS, Olsen JV. 2016. Analytic framework for peptidomics applied to large-scale neuropeptide identification. Nature Communications 7, 11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stes E, Laga M, Walton A, Samyn N, Timmerman E, De Smet I, Goormachtig S, Gevaert K. 2014. A COFRADIC protocol to study protein ubiquitination. Journal of Proteome Research 13, 3107–3113. [DOI] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL. 2005. Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nature Genetics 37, 1130–1134. [DOI] [PubMed] [Google Scholar]
- Tristan C, Shahani N, Sedlak TW, Sawa A. 2011. The diverse functions of GAPDH: views from different subcellular compartments. Cellular Signalling 23, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatsiani L, Timmerman E, De Bock PJ, et al. 2013. The Arabidopsis METACASPASE9 degradome. The Plant Cell 25, 2831–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen H, Overmyer K, Keinänen M, Kollist H, Kangasjärvi J. 2004. Mutual antagonism of ethylene and jasmonic acid regulates ozone-induced spreading cell death in Arabidopsis. The Plant Journal 39, 59–69. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Wallach D, Kang TB, Kovalenko A. 2014. Concepts of tissue injury and cell death in inflammation: a historical perspective. Nature Reviews. Immunology 14, 51–59. [DOI] [PubMed] [Google Scholar]
- van Doorn WG. 2011. Classes of programmed cell death in plants, compared to those in animals. Journal of Experimental Botany 62, 4749–4761. [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Beers EP, Dangl JL, et al. 2011. Morphological classification of plant cell deaths. Cell Death and Differentiation 18, 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K, Vandenberghe I, Van Beeumen J, Inzé D, Van Breusegem F. 2004. Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. The Journal of Biological Chemistry 279, 45329–45336. [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, del-Toro N, et al. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Research 44, D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M, Brosché M, Kollist H, Kangasjärvi J. 2009. Arabidopsis GRI is involved in the regulation of cell death induced by extracellular ROS. Proceedings of the National Academy of Sciences, USA 106, 5412–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M, Vainonen JP, Stael S, et al. 2015. GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. The EMBO Journal 34, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Vavylonis D, Durachko DM, Cosgrove DJ. 2017. Nanoscale movements of cellulose microfibrils in primary cell walls. Nature Plants 3, 17056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Sanchez-Lopez E, Karin M. 2016. Autophagy, inflammation, and immunity: a troika governing cancer and its treatment. Cell 166, 288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.