Abstract
Cathepsin A (CTSA) is a lysosomal protease that is abnormally expressed in various types of cancer; however, the function of CTSA in lung adenocarcinoma (LUAD) is unknown. The aim of the present study was to investigate the role of CTSA during LUAD development in vitro. The Cancer Genome Atlas (TCGA) database was used to analyze the expression of CTSA mRNA in LUAD tissues. CTSA was significantly upregulated in LUAD tissues compared with normal lung tissues. To explore the effect of CTSA on LUAD in vitro, LUAD A549 cells were transfected with CTSA small interfering RNA and the hallmarks of tumorigenesis were investigated using cell proliferation, cell cycle, wound healing, invasion and western blot assays. Following CTSA knockdown, proliferation of LUAD cells was decreased and an increased proportion of LUAD cells were arrested at the G0/G1 phase, with altered expression of critical cell cycle and proliferative marker proteins, including p53, p21 and proliferating cell nuclear antigen. Moreover, CTSA knockdown decreased the migration and invasion of A549 cells, as determined by wound healing, invasion, and western blotting assays. The expression levels of key proteins involved in epithelial-mesenchymal transition were analyzed by western blotting. CTSA knockdown enhanced the expression of E-cadherin, but decreased the expression of N-cadherin and β-catenin in A549 cells. To the best of our knowledge, the present study suggested for the first time it has been identified that CTSA may serve as a tumor promoter in LUAD, enhancing the malignant progression of LUAD cells by promoting cell proliferation, migration and invasion. The results suggested that CTSA may serve as a novel therapeutic target for LUAD.
Keywords: cathepsin A, lung adenocarcinoma, epithelial-mesenchymal transition, cell cycle, cell proliferation, cell migration, cell invasion
Introduction
Lung cancer is a malignant cancer, which demonstrates the highest rates of morbidity and mortality worldwide (1); the most common histological subtype is adenocarcinoma (2). Although the diagnosis and treatment of lung cancer has progressed in recent decades, the survival rate has not significantly improved (3). A total of ~80% of patients with lung cancer are diagnosed during the advanced stages of the disease, when treatment strategies are no longer optimal (4). Tumor metastasis is the leading cause of death in lung adenocarcinoma (LUAD); therefore, further investigation into the molecular mechanisms underlying LUAD metastasis is required for the identification of potential therapeutic targets for the disease.
Cathepsins are the key acid hydrolases in lysosomes and the main effector enzymes of protein catabolism and autophagy (5,6). Cathepsin A (CTSA) is a serine protease member of the cathepsin family that displays carboxypeptidase, deaminase and esterase activity and regulates the function of bioactive peptides (7,8). CTSA protects β-galactosidase and neuraminidase proteins from lysosomal proteolysis by forming multienzyme complexes (9). CTSA deficiency can cause human lysosomal storage disease, known as galactosialidosis (10). A recent study demonstrated that CTSA is a key enzyme involved in the degradation of lysosome-associated membrane protein type 2a, which is an important effector of chaperone-mediated autophagy (11). Several studies have confirmed that cathepsin proteases regulate cancer progression and the therapeutic response (12–14). High CTSA expression is associated with various tumors (15–17); however, whether abnormal expression of CTSA is associated with LUAD development is not completely understood.
The Cancer Genome Atlas (TCGA) is a public-funded project, aimed at cataloguing and discovering the major human genome variations that induce cancerization (18). By analyzing TCGA database, differential expression data of CTSA in LUAD and normal tissues can be obtained, which provides a foundation for the investigation of the roles of CTSA during LUAD molecular pathogenesis.
In the present study, the aim was to investigate whether CTSA knockdown decreased proliferation, migration and invasion of A549 cells, and to assess the oncogenic potential of CTSA in LUAD.
Materials and methods
Data source and bioinformatics analysis
Human RNA-sequencing data from LUAD projects which included 515 patients with LUAD and 59 normal tissues were collected from TCGA database (portal.gdc.cancer.gov). The R software package (version 3.6.0; RStudio, Inc.) was used to analyze the expression of CTSA in normal and tumor samples.
Cell culture
The human lung cancer cell line A549 was obtained from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Cells were cultured in RPMI-1640 medium (Biological Industries) supplemented with 10% FBS (Biological Industries) at 37°C with 5% CO2 and 95% air.
Cell transfection
A total of 5×105 A549 cells/well were transfected with 50 nM small interfering (si)RNA-CTSA (5′-GCCUGCCACUCAAGCGGAUTT-3′) or siRNA-negative control (NC; non-targeting; 5′-UUCUCCGAACGUGUCACGUTT-3′; Shanghai GenePharma Co., Ltd.) using Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Following a 48-h incubation at 37°C, cells were harvested for subsequent experiments.
RNA isolation and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from A549 cells using the RNAiso Plus kit (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. RNA purity was assessed using a NanoPhotometer® spectrophotometer. Total RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. The following experimental conditions were used for reverse transcription: 42°C for 2 min, 37°C for 15 min and 85°C for 5 sec. Subsequently, qPCR was performed using an ABI 7500 Fast Real-Time PCR system (Thermo Fisher Scientific, Inc.) and the SYBR Premix Ex Taq™ II kit (Takara Biotechnology Co., Ltd), according to the manufacturer's protocol. The following primer pairs were used for qPCR: CTSA forward, 5′-GTCGCCCAGAGCAATTTTGAG-3′ and reverse, 5′-TCTCCCCGGTCAGGAAAAGTT-3′; and β-actin forward, 5′-CATGTACGTTGCTATCCAGGC-3′ and reverse, 5′-CTCCTTAATGTCACGCACGAT-3′. The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 30 sec; followed by 40 cycles of 95°C for 5 sec and 60°C for 34 sec. CTSA mRNA levels were quantified using the 2−ΔΔCq method and normalized to the internal reference gene β-actin.
Cell proliferation assay
The effect of CTSA knockdown on cell proliferation was assessed using a Cell Counting Kit-8 (CCK-8) assay (Biosharp Life Sciences), according to the manufacturer's protocol. A549 cells were harvested 48 h post-transfection, seeded (5×103 cells/well) into a 96-well microplate and incubated at 37°C for 24, 48 or 72 h. Subsequently, 10% CCK-8 solution (10 µl CCK-8 reagent; 90 µl RPMI 1640 medium) was added to each well for 2 h at 37°C. The optical density of each well was determined at a wavelength of 450 nm using an Elx808 microplate reader (BioTek Instruments, Inc.).
Cell cycle analysis
Transfected A549 cells (2×106 cells/well) were seeded into 6-well plates, incubated overnight and cultured in RPMI-1640 medium without serum for 12 h to synchronize the cell cycle. The cells were fixed with 70% ethanol for 12 h at 4°C, washed with 1 ml pre-cooled PBS and stained with propidium iodide and RNase A solution (25 µg/ml; Beijing 4A Biotech Co., Ltd.) for 30 min at 37°C in the dark. The DNA content of the cells, used as an indicator of the different phases of the cell cycle, was measured using a FACSCalibur flow cytometer (BD Biosciences; Becton, Dickinson and Company) with CellQuest Pro (version 5.2; BD Biosciences; Becton, Dickinson and Company) and ModFit LT (version 3.0; Verity Software House, Inc.) software.
Western blot analysis
Total protein was extracted from A549 cells using RIPA lysis buffer (Beyotime Institute of Biotechnology) supplemented with phenylmethylsulphonyl fluoride (Beyotime Institute of Biotechnology). Proteins were isolated by centrifugation at 12,000 × g for 15 min at 4°C. Total protein was quantified using a bicinchoninic acid assay (Beyotime Institute of Biotechnology). Subsequently, protein samples were incubated with Loading Buffer (Beyotime Institute of Biotechnology) at 100°C for 10 min. Equal amounts of protein (30 µg per lane) were separated via 10% SDS-PAGE for 2 h at a constant voltage (110 V), and subsequently transferred onto PVDF membranes (EMD Millipore). The membranes were blocked using TBST (20 mM Tris-Hcl, 150 mM NaCl pH 7.5 and 0.1% Tween-20) containing 5% skim milk at room temperature for 1–2 h. Subsequently, the membranes were incubated overnight at 4°C with the following primary antibodies: Anti-CTSA (cat. no. ab184553; 1:10,000; Abcam), anti-E-cadherin (cat. no. WL01482; 1:1,000; Wanleibio Co., Ltd.), anti-p53 (cat. no. WL01919; 1:1,000; Wanleibio Co., Ltd.), anti-p21 (cat. no. WL0362; 1:1,000; Wanleibio Co., Ltd.), anti-proliferating cell nuclear antigen (PCNA; cat. no. WL01482; 1:1,000; Wanleibio Co., Ltd.), anti-N-cadherin (cat. no. 4061S; 1:1,000; Cell Signaling Technology, Inc.), anti-β-catenin (cat. no. AF6266; 1:1,000; Affinity Biosciences) and anti-β-actin (cat. no. 60008-1-lg; 1:4,000; ProteinTech, Group, Inc.). Following primary incubation, the membranes were washed three times with TBST and incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G (IgG; cat. no. ZB-2301; 1:20,000; OriGene Technologies, Inc.) and HRP-conjugated goat anti-mouse IgG (cat. no. ZB-2305; 1:20,000; OriGene Technologies, Inc.) secondary antibodies at 25°C for 2 h. Protein bands were visualized using the Amersham Imager 600 enhanced chemiluminescence detection system (GE Healthcare Life Sciences) and quantified using Image J software (version 1.52; National Institutes of Health), with β-actin as the loading control.
Wound healing assay
The migratory ability of A549 cells was assessed using wound healing assays. At 48 h post-transfection, A549 cells were seeded into a 6-well plate and subsequently grown to 90% confluence. A 200 µl pipette tip was used to make a single scratch through the cell monolayer and PBS was used to remove debris. A549 cells were cultured in RPMI-1640 medium, supplemented with 2% FBS to maintain the adherence of the cells for 48 h. Cell migration was observed at 0, 24 and 48 h using a light microscope (magnification, ×100; Nikon Corporation) and quantified using ImageJ software (version 1.37; National Institutes of Health). The migration rate was defined as the percentage of wound closure.
Transwell assay
Transwell assays were used to assess the invasive ability of A549 cells. Prior to the assay, the 24-well Transwell plates (8-µM; Corning Life Sciences) were pre-coated with 40 µl Matrigel® (1.5 mg/ml; BD Biosciences; Becton, Dickinson and Company) overnight at 37°C. At 48 h post-transfection, A549 cells (5×104) were suspended in 200 µl RPMI-1640 medium without serum and seeded into the upper chambers of the Transwell plates. RPMI-1640 medium supplemented with 20% FBS (500 µl) was added to the lower chambers of the Transwell plates to act as a chemoattractant stimulus. Subsequently, the Transwell plates were incubated for 48 h at 37°C. Cells on the upper surface of the membrane were wiped off using cotton swabs and invading cells on the lower surface of the membrane were fixed with 4% paraformaldehyde for 30 min at room temperature, and stained with 0.1% crystal violet (Beijing Solarbio Science & Technology Co., Ltd.) for 15 min at room temperature. The invasive ability of A549 cells was calculated under a light microscope as the mean number of cells in four randomly selected fields of view (magnification, ×100).
Statistical analysis
Statistical analyses were performed using GraphPad Prism software (version 5.0; GraphPad Software, Inc.). Data are presented as the mean ± standard deviation. One-way ANOVA followed by the least significant distance post hoc test was used make comparisons. All experiments were repeated at least three times. P<0.05 was considered to indicate a statistically significant difference.
Results
CTSA expression in LUAD tissues
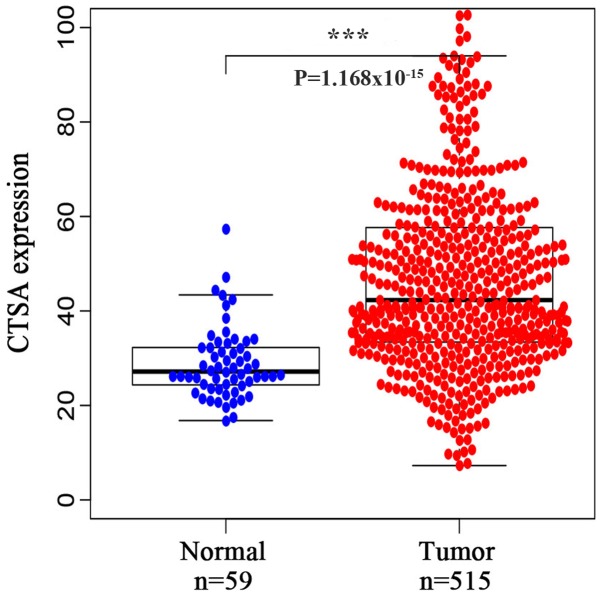
To investigate the expression of CTSA in LUAD, a bioinformatics analysis of data obtained from TCGA database was conducted. CTSA mRNA expression was significantly increased in LUAD tissues compared with normal lung tissues (P<0.001; Fig. 1).
Figure 1.
CTSA expression based on data obtained from TCGA database. CTSA expression was significantly increased in LUAD tissues compared with normal tissues. ***P<0.001 vs. normal lung tissues. CTSA, cathepsin A; TCGA, The Cancer Genome Atlas; LUAD, lung adenocarcinoma.
CTSA expression following CTSA knockdown
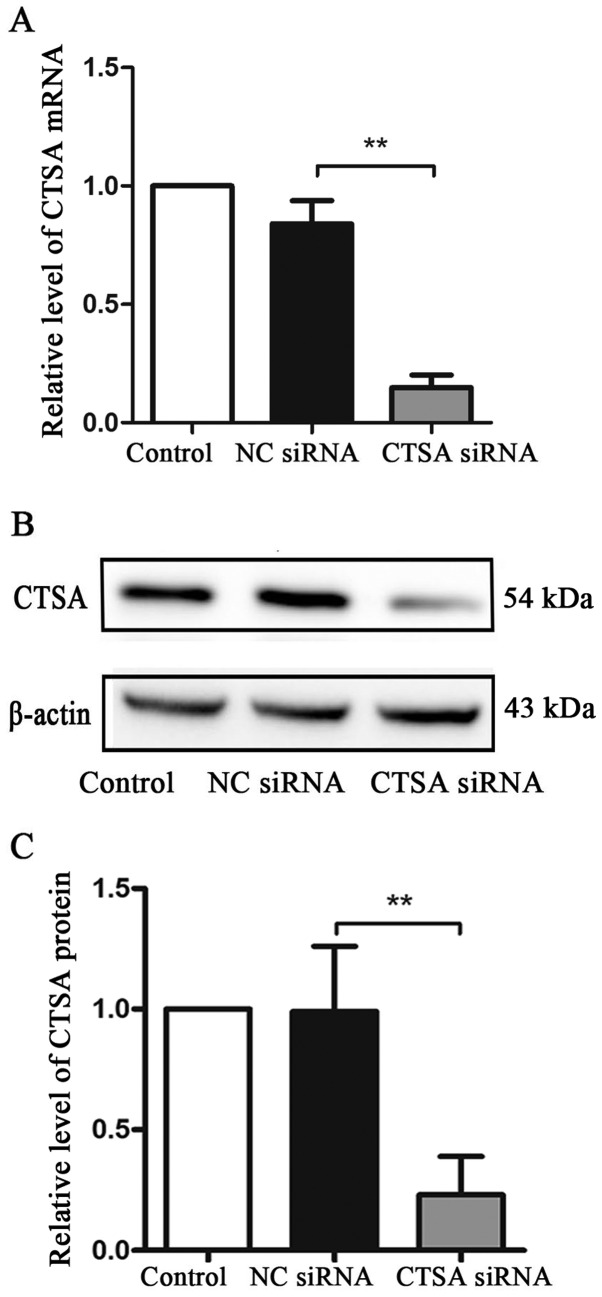
To investigate the effect of CTSA on the biological characteristics of A549 cells, CTSA siRNA was used to knockdown the expression of CTSA. Transfection efficiency was demonstrated using RT-qPCR and western blotting. In A549 cells, CTSA mRNA and protein expression levels were significantly decreased in CTSA knockdown cells compared with NC siRNA cells (P<0.01; Fig. 2A-C).
Figure 2.
CTSA mRNA and protein expression levels in transfected A549 cells. (A) CTSA mRNA expression levels were determined by reverse transcription-quantitative PCR. CTSA protein expression levels were (B) determined by western blotting and (C) quantified. **P<0.01. CTSA, cathepsin A; NC, negative control; siRNA, small interfering RNA.
CTSA knockdown decreases the proliferation of A549 cells
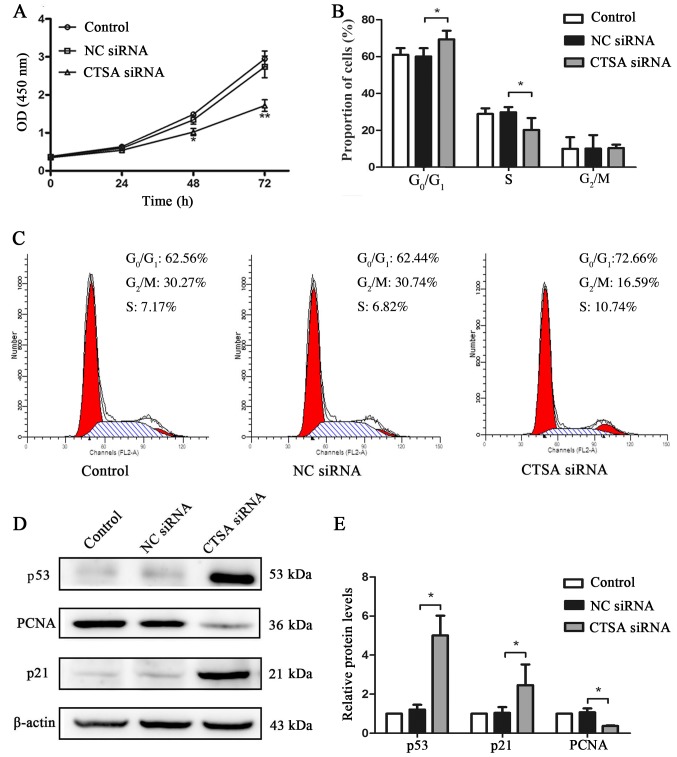
The proliferation of CTSA-knockdown cells was significantly decreased compared with the negative control cells, as assessed by the CCK-8 assay (P<0.01; Fig. 3A). CTSA knockdown in A549 cells resulted in a significant increase in the proportion of cells in the G0/G1 phase and a decrease in the proportion of cells in the S phase compared with the negative control cells (P<0.05; Fig. 3B and C). In addition, the expression levels of cell proliferation and cell cycle-associated proteins were detected by western blotting. The CTSA siRNA group displayed significantly decreased expression levels of PCNA and significantly increased expression levels of p53 and p21 compared with the NC siRNA group (P<0.01; Fig. 3D and E). The results indicated that CTSA knockdown decreased the proliferation of LUAD cells.
Figure 3.
Effects of CTSA knockdown on the proliferation, cell cycle and protein expression of A549 cells. (A) Proliferation of A549 cells transfected with CTSA siRNA and NC siRNA was measured using the Cell Counting Kit-8 assay. **P<0.01 vs. the NC siRNA group. (B) The proportion of transfected A549 cells in each stage of the cell cycle was detected by flow cytometry. *P<0.05, as indicated. (C) Representative plots of the flow cytometry analysis of cell cycle distribution in transfected A549 cells. Protein expression levels of p21, p53 and PCNA in transfected A549 cells were (D) determined by western blotting and (E) quantified. *P<0.05. CTSA, cathepsin A; siRNA, small interfering RNA; NC, negative control; OD, optical density; PCNA, proliferating cell nuclear antigen.
CTSA knockdown decreases the migration, invasion and EMT of LUAD cells
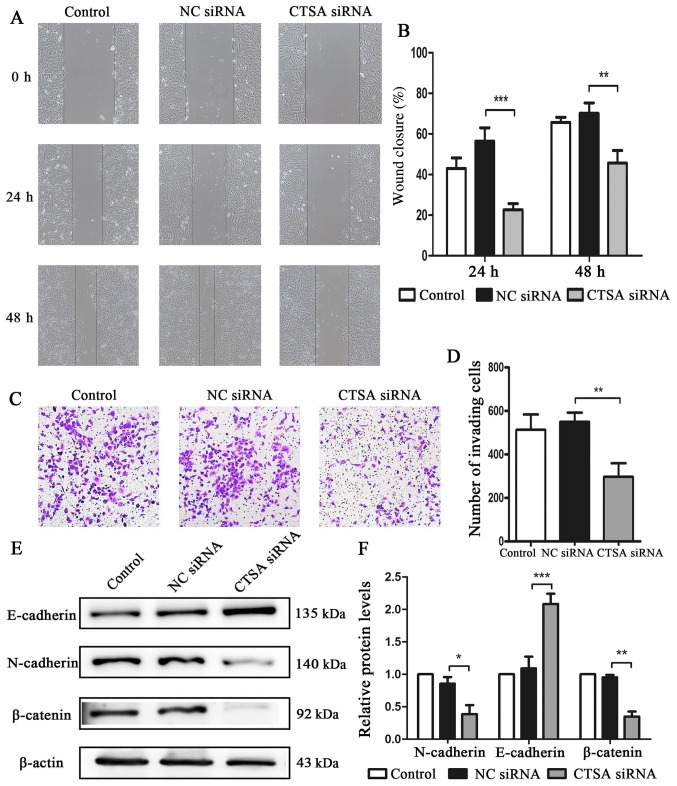
The migratory and invasive abilities of CTSA-knockdown cells were investigated using wound healing and invasion assays. The siRNA-CTSA group displayed a significant decrease in migratory (P<0.01; Fig. 4A and B) and invasive (P<0.01; Fig. 4C and D) abilities compared with the NC siRNA group. The expression of the main proteins involved in EMT was assessed to investigate the effects of CTSA knockdown on invasion and migration. The CTSA siRNA group displayed increased E-cadherin expression (P<0.001), but decreased levels of N-cadherin (P<0.05) and β-catenin (P<0.01) expression compared with the NC siRNA group (Fig. 4E and F). The results suggested that CTSA knockdown decreased the migration and invasion of LUAD cells.
Figure 4.
Effects of CTSA knockdown on the migration, invasion and EMT of A549 cells. Cell migration was (A) determined by wound healing assays and (B) quantified (magnification, ×100). Cell invasion was (C) determined by Transwell invasion assays and (D) quantified (magnification, ×100). Protein expression levels of E-cadherin, N-cadherin and β-catenin were (E) determined by western blotting and (F) quantified. *P<0.05, **P<0.01 and ***P<0.001, as indicated. CTSA, cathepsin A; EMT, epithelial-mesenchymal transition; NC, negative control; siRNA, small interfering RNA.
Discussion
Lung cancer is one of the most common types of cancer and is a major cause of cancer-related mortality. Proliferation and invasion play important roles during tumor development, and the ability to inhibit tumor growth and metastasis is key to successful treatment (19). Although various therapeutic targets for lung cancer have been identified, the survival rate has not significantly improved (20). However, there are only a few targets that can inhibit multiple tumor progression processes; therefore, it is critical to identify further pleiotropic therapeutic targets for LUAD.
Cathepsins, as lysosomal proteases, participate in multiple processes associated with cancer, including protein degradation, autophagy, growth factor receptor recycling and lysosome-mediated cell death. Abnormal expression of cathepsins has been reported to be associated with a variety of different types of cancer (8,21–23). For example, previous studies have reported that abnormal expression of cathepsins, including cathepsin K, L and S, is observed in lung cancer (24–26), which suggests that variations in cathepsin expression may play a role in lung cancer. A number of studies have indicated that CTSA is involved in breast, colorectal and melanocyte cancer, and is also associated with tumorigenesis, malignant progression and poor patient prognosis (15–17). However, the role of CTSA in LUAD has not been reported. In the present study, the bioinformatics analysis of TCGA database suggested that the expression of CTSA was upregulated in LUAD tissues compared with normal tissues, suggesting that high CTSA expression was associated with LUAD.
In various types of cancer, cathepsins are highly upregulated and their increased expression correlates with more aggressive tumor types and poor patient prognosis (8,27–33). Cathepsins contribute to the degradation of the basement membrane that confines the tumor to promote invasion and tumor metastasis (34). The expression levels of CTSA are higher in metastatic melanoma compared with primary melanoma and are associated with poor prognosis (17). Similarly, higher CTSA expression is associated with aggressive tumor types and poor outcomes in breast and colorectal cancer (CRC), and reducing CTSA expression inhibits the migration and invasion of CRC cells (15,16).
CTSA knockdown led to a reduction in the migration and invasion of LUAD cells. EMT, which is a cellular process that involves the loss of polarization and intercellular connections in epithelial cells, plays an important role in tumor metastasis and recurrence (35). A key characteristic of EMT is the loss of E-cadherin expression and the gain of N-cadherin expression in the cell membrane, which is known as the E-to-N-cadherin switch (36). E-cadherin has been identified as a substrate for CTSB, CTSL and CTSS in tumors (37). CTSA is highly expressed in A549 cells; therefore, overexpression of CTSA in A549 cells was not performed in the present study. However, CTSA knockdown was performed to investigate the relationship between CTSA and EMT in LUAD cells. The results suggested that CTSA knockdown increased the expression of the epithelial marker E-cadherin and decreased the expression of N-cadherin and β-catenin in A549 cells.
The catabolism of proteins is one of the major functions of proteases. Cathepsin deficiencies result in autophagosome accumulation, which indicates impaired catabolism of the cargo and decreased autophagic flux (6). Increased lysosomal biogenesis and cathepsin expression in cancer cells supports cell survival (38). Furthermore, increased catabolic activity is essential to provide the necessary nutrients to maintain cell proliferation. Therefore, it was hypothesized that the loss of cathepsin may inhibit cell proliferation by impairing the catabolic activity and the lack of essential nutrients. In the present study, CTSA knockdown decreased the proliferation and increased cell cycle arrest at the G0/G1 phase in LUAD cells. Further experiments suggested that CTSA knockdown altered the expression of critical cell cycle-associated proteins, including p53, p21 and the proliferative marker PCNA. p53 is a transcription factor that initiates cell cycle arrest, cell senescence or apoptosis to regulate tumor progression. p21 is involved in p53-mediated cycle arrest, which inhibits cyclin E-CDK2 activity (39).
To the best of our knowledge, the present study suggested for the first time that CTSA may serve as an oncogene in LUAD. CTSA knockdown decreased the proliferation, migration and invasion of A549 cells, and altered the expression of key cell cycle regulators. However, in order to maintain the cells in a good adherence state, 2% FBS was used in the wound healing assay, which is a limitation of the present study in terms of assessing the migratory ability. CTSA also displayed pleiotropic effects in A549 cells, which suggested that CTSA may serve as a vital effector during tumor development, indicating that targeting CTSA may allow multiple tumorigenic processes to be inhibited via a single therapeutic target. The results of the present study suggested that CTSA may serve as a potential therapeutic target for LUAD; however, further investigation into the mechanisms underlying the role of CTSA in LUAD, including in vitro and in vivo experiments, is required.
Acknowledgements
Not applicable.
Funding
The present study was supported by The China Medical University Discipline Enhancement Program Support Project.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request. The datasets generated and/or analyzed during the current study are available in the TCGA repository, (http://cancergenome.nih.gov).
Authors' contributions
JL, BH and XZ conceived and designed the study. BH performed the experiments and wrote the manuscript. BH and XZ analyzed the data and critically revised the manuscript. JL and XZ supervised the study and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network, corp-author. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janssen-Heijnen ML, van Erning FN, De Ruysscher DK, Coebergh JW, Groen HJ. Variation in causes of death in patients with non-small cell lung cancer according to stage and time since diagnosis. Ann Oncol. 2015;26:902–907. doi: 10.1093/annonc/mdv061. [DOI] [PubMed] [Google Scholar]
- 4.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 5.Pan L, Li Y, Jia L, Qin Y, Qi G, Cheng J, Qi Y, Li H, Du J. Cathepsin S deficiency results in abnormal accumulation of autophagosomes in macrophages and enhances Ang II-induced cardiac inflammation. PLoS One. 2012;7:e35315. doi: 10.1371/journal.pone.0035315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennemärker J, Lohmüller T, Müller S, Aguilar SV, Tobin DJ, Peters C, Reinheckel T. Impaired turnover of autophagolysosomes in cathepsin L deficiency. Biol Chem. 2010;391:913–922. doi: 10.1515/bc.2010.097. [DOI] [PubMed] [Google Scholar]
- 7.Drake MT, Clarke BL, Oursler MJ, Khosla S. Cathepsin K inhibitors for osteoporosis: Biology, potential clinical utility, and lessons learned. Endocr Rev. 2017;38:325–350. doi: 10.1210/er.2015-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olson OC, Joyce JA. Cysteine cathepsin proteases: Regulators of cancer progression and therapeutic response. Nat Rev Cancer. 2015;15:712–729. doi: 10.1038/nrc4027. [DOI] [PubMed] [Google Scholar]
- 9.Akkari L, Gocheva V, Quick ML, Kester JC, Spencer AK, Garfall AL, Bowman RL, Joyce JA. Combined deletion of cathepsin protease family members reveals compensatory mechanisms in cancer. Genes Dev. 2016;30:220–232. doi: 10.1101/gad.270439.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolli N, Garman SC. Proteolytic activation of human cathepsin A. J Biol Chem. 2014;289:11592–11600. doi: 10.1074/jbc.M113.524280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.You Y, Li WZ, Zhang S, Hu B, Li YX, Li HD, Tang HH, Li QW, Guan YY, Liu LX, et al. SNX10 mediates alcohol-induced liver injury and steatosis by regulating the activation of chaperone-mediated autophagy. J Hepatol. 2018;69:129–141. doi: 10.1016/j.jhep.2018.01.038. [DOI] [PubMed] [Google Scholar]
- 12.Verbovšek U, Van Noorden CJ, Lah TT. Complexity of cancer protease biology: Cathepsin K expression and function in cancer progression. Semin Cancer Biol. 2015;35:71–84. doi: 10.1016/j.semcancer.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Sudhan DR, Siemann DW. Cathepsin L targeting in cancer treatment. Pharmacol Ther. 2015;155:105–116. doi: 10.1016/j.pharmthera.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan L, Sheng L, He W, Zou C, Hu B, Liu J, Ge W, Liu Y, Wang J, Ma E. Discovery of novel cathepsin inhibitors with potent anti-metastatic effects in breast cancer cells. Bioorg Chem. 2018;81:672–680. doi: 10.1016/j.bioorg.2018.09.029. [DOI] [PubMed] [Google Scholar]
- 15.Ni S, Weng W, Xu M, Wang Q, Tan C, Sun H, Wang L, Huang D, Du X, Sheng W. miR-106b-5p inhibits the invasion and metastasis of colorectal cancer by targeting CTSA. Onco Targets Ther. 2018;11:3835–3845. doi: 10.2147/OTT.S172887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toss MS, Miligy IM, Haj-Ahmad R, Gorringe KL, AlKawaz A, Mittal K, Ellis IO, Green AR, Rakha EA. The prognostic significance of lysosomal protective protein (cathepsin A) in breast ductal carcinoma in situ. Histopathology. 2019;74:1025–1035. doi: 10.1111/his.13835. [DOI] [PubMed] [Google Scholar]
- 17.Kozlowski L, Wojtukiewicz MZ, Ostrowska H. Cathepsin A activity in primary and metastatic human melanocytic tumors. Arch Dermatol Res. 2000;292:68–71. doi: 10.1007/s004030050012. [DOI] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research Network, corp-author. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung JJ, Yeh YC, Jeng WJ, Wu KJ, Huang BS, Wu YC, Chou TY, Hsu WH. Predictive value of the international association for the study of lung cancer/American thoracic society/European respiratory society classification of lung adenocarcinoma in tumor recurrence and patient survival. J Clin Oncol. 2014;32:2357–2364. doi: 10.1200/JCO.2013.50.1049. [DOI] [PubMed] [Google Scholar]
- 21.Reinheckel T, Hagemann S, Dollwet-Mack S, Martinez E, Lohmuller T, Zlatkovic G, Tobin DJ, Maas-Szabowski N, Peters C. The lysosomal cysteine protease cathepsin L regulates keratinocyte proliferation by control of growth factor recycling. J Cell Sci. 2005;118:3387–3395. doi: 10.1242/jcs.02469. [DOI] [PubMed] [Google Scholar]
- 22.Platt MO, Shockey WA. Endothelial cells and cathepsins: Biochemical and biomechanical regulation. Biochimie. 2016;122:314–323. doi: 10.1016/j.biochi.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brojatsch J, Lima H, Jr, Palliser D, Jacobson LS, Muehlbauer SM, Furtado R, Goldman DL, Lisanti MP, Chandran K. Distinct cathepsins control necrotic cell death mediated by pyroptosis inducers and lysosome-destabilizing agents. Cell Cycle. 2015;14:964–972. doi: 10.4161/15384101.2014.991194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Zhao Y, Xiong Y, Wang W, Fei Y, Tan C, Liang Z. K-ras mutation promotes ionizing radiation-induced invasion and migration of lung cancer in part via the Cathepsin L/CUX1 pathway. Exp Cell Res. 2018;362:424–435. doi: 10.1016/j.yexcr.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Willumsen N, Bager CL, Leeming DJ, Bay-Jensen AC, Karsdal MA. Nidogen-1 degraded by cathepsin S can be quantified in serum and is associated with non-small cell lung cancer. Neoplasia. 2017;19:271–278. doi: 10.1016/j.neo.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai JY, Lee MJ, Dah-Tsyr Chang M, Huang H. The effect of catalase on migration and invasion of lung cancer cells by regulating the activities of cathepsin S, L, and K. Exp Cell Res. 2014;323:28–40. doi: 10.1016/j.yexcr.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Jechorek D, Votapek J, Meyer F, Kandulski A, Roessner A, Franke S. Characterization of cathepsin X in colorectal cancer development and progression. Pathol Res Pract. 2014;210:822–829. doi: 10.1016/j.prp.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, Wang S, Wang Q, Yang Z, Pan Z, Li L. Overexpression of cysteine cathepsin L is a marker of invasion and metastasis in ovarian cancer. Oncol Rep. 2014;31:1334–1342. doi: 10.3892/or.2014.2967. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita K, Iwatake M, Okamoto K, Yamada SI, Umeda M, Tsukuba T. Cathepsin K modulates invasion, migration and adhesion of oral squamous cell carcinomas in vitro. Oral Dis. 2017;23:518–525. doi: 10.1111/odi.12643. [DOI] [PubMed] [Google Scholar]
- 30.Aggarwal N, Sloane BF. Cathepsin B: Multiple roles in cancer. Proteomics Clin Appl. 2014;8:427–437. doi: 10.1002/prca.201300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schweiger A, Staib A, Werle B, Krasovec M, Lah TT, Ebert W, Turk V, Kos J. Cysteine proteinase cathepsin H in tumours and sera of lung cancer patients: Relation to prognosis and cigarette smoking. Br J Cancer. 2000;82:782–788. doi: 10.1054/bjoc.1999.0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruan J, Zheng H, Fu W, Zhao P, Su N, Luo R. Increased expression of cathepsin L: A novel independent prognostic marker of worse outcome in hepatocellular carcinoma patients. PLoS One. 2014;9:e112136. doi: 10.1371/journal.pone.0112136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han ML, Zhao YF, Tan CH, Xiong YJ, Wang WJ, Wu F, Fei Y, Wang L, Liang ZQ. Cathepsin L upregulation-induced EMT phenotype is associated with the acquisition of cisplatin or paclitaxel resistance in A549 cells. Acta Pharmacol Sin. 2016;37:1606–1622. doi: 10.1038/aps.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turk V, Turk B, Guncar G, Turk D, Kos J. Lysosomal cathepsins: Structure, role in antigen processing and presentation, and cancer. Adv Enzyme Regul. 2002;42:285–303. doi: 10.1016/S0065-2571(01)00034-6. [DOI] [PubMed] [Google Scholar]
- 35.Pastushenko I, Blanpain C. EMT Transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gocheva V, Zeng W, Ke D, Klimstra D, Reinheckel T, Peters C, Hanahan D, Joyce JA. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006;20:543–556. doi: 10.1101/gad.1407406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kallunki T, Olsen OD, Jäättelä M. Cancer-associated lysosomal changes: Friends or foes? Oncogene. 2013;32:1995–2004. doi: 10.1038/onc.2012.292. [DOI] [PubMed] [Google Scholar]
- 39.Harris SL, Levine AJ. The p53 pathway: Positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request. The datasets generated and/or analyzed during the current study are available in the TCGA repository, (http://cancergenome.nih.gov).