Abstract
Cancer cells with cancer stem cell (CSC) properties initiate both primary tumor formation and metastases at distant sites. Acquisition of CSC properties is highly associated with epigenetic alterations, including those mediated by microRNAs (miRNAs). We have previously established the breast cancer patient‐derived tumor xenograft (PDX) mouse model in which CSC marker CD44+ cancer cells formed spontaneous microscopic metastases in the liver. In this PDX mouse, we found that the expression levels of 3 miRNAs (miR‐25, miR‐93, and miR‐106b) in the miR‐106b‐25 cluster were much lower in the CD44+ human cancer cells metastasized to the liver than those at the primary site. Constitutive overexpression of miR‐93 suppressed invasive ability and 3D‐organoid formation capacity of breast cancer cells in vitro and significantly suppressed their metastatic ability to the liver in vivo. Wiskott‐Aldrich syndrome protein family member 3 (WASF3), a regulator of both cytoskeleton remodeling and CSC properties, was identified as a functional target of miR‐93: overexpression of miR‐93 reduced the protein level of WASF3 in breast cancer cells and WASF3 rescued the miR‐93‐mediated suppression of breast cancer cell invasion. These findings suggest that miR‐93 functions as a metastasis suppressor by suppressing both invasion ability and CSC properties in breast cancers.
Keywords: breast cancer, cancer stem cell, metastasis, miR‐93, WASF3
Multiplex microRNA (miRNA) expression analyses of primary and metastasized cancer cells in breast cancer patient‐derived tumor xenograft mouse revealed the downregulation of the miR‐106b‐25 cluster miRNAs in the CD44+ cancer cells metastasized to the liver. Among them, miR‐93 suppressed invasion and organoid formation ability of breast cancer cells, and WASF3 was identified as a functional target of miR‐93. Finally, we found that overexpression of miR‐93 suppressed the metastasis of breast cancer cells in vivo.
1. INTRODUCTION
Advanced stage, metastatic breast cancers are difficult to cure; nearly 90% of these breast cancer‐associated deaths were due to metastasis, a clinically incurable disease. 1 The metastatic cascade is composed of highly complex multistep processes, including metabolic reprogramming, the ability to enter and exit dormancy, resistance to apoptosis, immune evasion, and interaction with other tumor and stromal cells. 2
MicroRNAs (miRNAs) contribute to the post‐transcriptional regulation of mRNAs. MicroRNAs regulate various mechanisms underlying metastasis, such as epithelial‐mesenchymal transition (EMT), exosome secretion, cell survival and invasion. For example, metastasis of breast cancer is promoted by miR‐10b, miR‐9, miR‐206, miR‐335, miR‐126, and miR‐19a. 3 MicroRNA‐10b, whose expression level is elevated in metastasis‐positive breast cancer patients, targets tumor suppressors (eg, TBX5 and PTEN) and metastasis suppressors (eg, HOXD10) 4 , 5 ; miR‐9 targets CDH1 and leukemia inhibitory factor receptor that is downregulated in human breast cancer and functions a marker for survival outcomes. 6 , 7 MicroRNA‐19a from astrocyte‐derived exosomes targets PTEN and promotes brain metastasis. 8
Wiskott‐Aldrich syndrome protein family member 3 (WASF3) is an actin cytoskeleton remodeling protein, is highly expressed in advanced stages of breast cancer, and promotes tumor cell invasion and metastasis, especially through its phosphorylation by human epidermal growth factor receptor 2 (HER2)/ERBB2 signaling. 9 WASF3 protein regulates actin cytoskeleton dynamics through activation of the Arp2/3 complex and binds to actin through a C‐terminal verprolin homology domain. It is involved in various aspects of cancers, such as metastasis, tumor growth, cell cycle progression, and drug resistance. Indeed, metastasis‐promoting roles of WASF3 in breast cancer are revealed using a Wasf3 null/polyoma middle‐T oncogene mouse model. 10 WASF3 downregulates miR‐200 family miRNAs, suppressors of EMT, during tumor progression, 11 , 12 , 13 suggesting that WASF3 and miR‐200 play a key role in controlling the invasion‐metastasis cascade of cancer cells. WASF3 is one of the targets of miRNAs, such as miR‐7 and miR‐217, that inhibit the motility and/or metastatic potential of cancer cells. 14 , 15
Cancer stem cells (CSCs) are subpopulation of the cells that retain tumorigenic capacity following serial transplantation and, at the same time, are able to sustain the formation of tumors that recreate the cellular diversity of the parent lesions from which they have been originally isolated. 16 Furthermore, highly tumorigenic properties of CSCs are associated with metastatic progression, especially at the initial steps of metastases. 17 In the specific case of human breast cancers, the subset of malignant cells endowed with CSC properties is enriched among cells defined by the CD44+/CD24low/neg phenotype. 16 , 18 , 19 We and others have shown that in epithelial malignancies such as breast cancer, self‐renewal ability of malignant cells is negatively regulated by miR‐200c, which suppresses the expression of BMI1. 13 , 18 In addition, miRNAs, such as let‐7, miR‐142, miR‐200c, and miR‐221, epigenetically regulate the properties of CSCs of patient‐derived tumor xenograft (PDX) cells by targeting APC, ZEB1, and RNA‐binding protein QKI. 16 , 20 , 21 , 22
In this study, we analyzed the metastasized CD44+ breast cancer cells in breast cancer PDX mice to identify intrinsic factors that were associated with cancer cell metastasis. Multiplex miRNA expression analysis of the primary and metastasized cancer cells in the PDX mouse revealed the downregulation of the miR‐106b‐25 cluster miRNAs in the CD44+ cancer cells metastasized to the liver. Among them, miR‐93 suppressed invasion ability and organoid formation capacity of breast cancer cells. We then identified that WASF3 was a functional target of miR‐93. Finally, we found that overexpression of miR‐93 suppressed the metastatic abilities of breast cancer cells in vivo.
2. MATERIALS AND METHODS
2.1. Ethics statements
Human primary breast cancers were obtained from patients admitted to the Division of Breast and Endocrine Surgery of Kobe University Hospital. The research was preapproved by Kobe University’s Institutional Review Board (permission numbers 1299 and 1481) and was carried out in accordance with recognized ethical guidelines. All patients included in the study provided written informed consent. Animal experiments were undertaken with the approval of Kobe University’s Animal Care and Use Committee (permission numbers P100905 and P150802) and carried out according to the Kobe University Animal Experiment Regulation.
2.2. Flow cytometry
Primary tumor specimens, xenograft tumors, and the liver of the PDX mice were dissociated using collagenase III (Worthington) and analyzed as previously described (Figure S1). 22 Dissociated cells were stained with mAbs conjugated to fluorescent dyes. Antibodies used in this study are anti‐human CD44‐allophycocyanin (APC, clone IM7, 1:20; BioLegend), anti‐human HLA A, B, C‐Alexa488 (clone W6/32, 1:20; BioLegend), anti‐mouse H‐2Kd/H2‐Dd‐biotin (clone 34‐1‐2S; eBioscience), and anti‐mouse Cd45‐biotin (clone 30‐F11; BD Biosciences) Abs.
2.3. Analysis of miRNA expression by multiplex semiquantitative real‐time PCR
RNA was extracted from 100 cells sorted from breast cancer PDX and directly collected into TRIzol (Invitrogen). The expression level of 754 miRNAs was measured by multiplex semiquantitative real‐time PCR (TaqMan Array Human MicroRNA A+B Cards Set version 3.0 with Megaplex RT Primers, Human Pool Set version 3.0; Thermo Fisher Scientific) as previously described. 22 Results were normalized to RNU48 small nuclear RNA (snRNA).
2.4. Semiquantitative RT‐PCR
The expression levels of miR‐93 were analyzed by semiquantitative RT‐PCR (RT‐qPCR) as described previously. 22 Briefly, RNA was extracted from a minimum of 100 cells from each population using TRIzol (Thermo Fisher Scientific). Reverse transcription was carried out using the SuperScript‐III RT kit (Thermo Fisher Scientific). In each sample, the expression level of miR‐93 was measured by RT‐qPCR using a specific primer set for the mature miR‐93 sequence (Thermo Fisher Scientific), and was normalized to that of RNU48 snRNA. To analyze the expression levels of mRNAs, RNA samples were reverse transcribed using a High Capacity cDNA Reverse Transcription Kit (Life Technologies). Semiquantitative PCR was undertaken as described previously. 23 The sequences of primers used are presented in Table S1. Data were normalized to the amount of Gapdh cDNA used as an endogenous control.
2.5. Cell lines
All cell lines used in this study were obtained from the ATCC (http://www.atcc.org) and include: MDA‐MB‐231, T‐47D, and MCF7 human breast cancer cells (ATCC catalog: HTB‐26, HTB‐133, and HTB‐22, respectively) and HEK293 human embryonic kidney cells (ATCC catalog: CRL‐1573). All cell lines were cultured in RPMI‐1640 (Sigma‐Aldrich) containing 10% FBS, penicillin (100 U/mL), and streptomycin (100 mg/mL; Nacalai). Early passage cells were used in all experiments.
2.6. Lentivirus production
The sequence of precursor miR‐93 (mature miR‐93 and its 5′‐ and 3′‐ flanking regions) and the full‐length coding region of the WASF3 mRNA (NM_006646.6 [GenBank]) were amplified by PCR and cloned into the pEIZ‐HIV‐ZsGreen or mCherry lentivirus vector (Addgene: #18121) or the pLentiLox3.7‐EF1α‐mCherry vector, a derivative of pLentiLox3.7 (Addgene: #11795), respectively. 18 The lentivirus vectors encoding for the anti‐miR‐93 construct (miRZip‐93) and a nontargeting control (negative control) were purchased from System Biosciences. Lentiviruses were produced as previously described. 24 Breast cancer cells were infected with lentivirus constructs at a MOI of 5.
2.7. Transwell cell invasion assay
Breast cancer cells were transfected with the miR‐93 mimic (Bioneer), miR‐93‐5p inhibitor (Ambion, Thermo Fisher Scientific) or corresponding negative controls using the Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s protocol. Transwell cell invasion assays were undertaken using a 24‐well transwell inserts with 8‐µm pore size (Corning). The upper surface of a filter membrane was coated with 30 μL Matrigel (Corning). Five thousand cells in DMEM without FBS were added to the upper compartment of the chamber; DMEM containing 10% FBS was added to the bottom chamber. After incubation at 37°C for 24 h, cells on the upper side of the membrane were removed using a cotton swab. The cells that invaded into the bottom chamber were fixed in 10% formaldehyde and stained in 1% crystal violet. The number of cells in 5 random fields per membrane were counted under a microscope.
2.8. Organoid assays
Cells were infected with lentivirus constructs for the expression of miR‐93, anti‐miR‐93, or corresponding control lentivirus constructs, and seeded on Matrigel (Corning) in 96‐well plates (3 × 103 cells/well), and cultured at 37°C with 5% CO2, as previously described. 25 The number of organoids larger than 100 μm in diameter was counted 10 days after seeding.
2.9. Cloning and mutagenesis of WASF3 3′‐UTR
The fragments of the WASF3 3′‐UTR (2473‐2944 [472 bp] and 3664‐4016 [353 bp] of NM_006646.6 [GenBank]) was amplified by PCR and cloned into the pGL3‐MC vector, at the 3′‐end of the firefly luciferase gene. 18 Mutations in the putative miR‐93 target sequence within the WASF3 3′‐UTR were introduced using a PrimeSTAR Mutagenesis Basal Kit (Takara Bio).
2.10. Luciferase reporter assays
Cells were cotransfected with: (i) a pGL3‐MC luciferase expression construct 18 ; (ii) the pRL‐TK Renilla luciferase vector (Promega); and (iii) a pEIZ expression plasmid containing either miR‐93 or an empty backbone, using Lipofectamine 3000 (Thermo Fisher Scientific). Luciferase activity was quantified and normalized to Renilla luciferase activity, using the Dual‐Luciferase Reporter Assay System (Promega).
2.11. Western blot analysis
Cells were lysed using SDS sample buffer. Cell lysates were separated by electrophoresis on a SDS‐10.5% polyacrylamide gel and transferred to a PVDF membrane. After blocking, filters were incubated with either an anti‐WASF3 (#2806, 1:1000; Cell Signaling Technology) or an anti‐β‐actin mAb (#017‐24573, 1:20 000; Wako) at 4°C overnight and then with a goat anti‐mouse IgG‐ HRP conjugate (#7074, 1:10 000; Cell Signaling Technology). Protein bands were detected using the Chemi‐Lumi One L bioluminescent assay (Nacalai).
2.12. Hepatic metastasis model by splenic injection of tumor cells and competitive transplantation assays
Skin and peritoneum at the left subcostal region of NOD/SCID/IL2Rγ−/− (NSG) mice (Charles River) was incised, and the spleen was exposed. Breast cancer cells were injected slowly into the spleen. A whitening of the spleen and blood vessels were observed upon injection. 26 Twenty to 30 days following the surgery, the mice were perfused with PBS and the liver was harvested. The part of the liver was fixed for histological examination and the rest of the liver was cut into pieces for dissociation. In the competitive transplantation assays, equal numbers of MDA‐MB‐231‐ZsGreen cells overexpressing miR‐93 (miR‐93 ZsGreen) and MDA‐MB‐231 mCherry competitor were mixed 1:1 and injected into the spleen.
2.13. Statistical analysis
Data are presented as means ± SD. Comparisons between continuous data normally distributed with equal variance or unequal variances between groups were undertaken using unpaired 2‐tailed Student’s t tests. Sample sizes, statistical tests, and P values are indicated in the figure or figure legends. All P values were 2‐sided and P values less than .05 were deemed statistically significant. Asterisks denote P value significance.
A description of the methods used in the experiments for supplementary figures is provided in Appendix S1.
3. RESULTS
3.1. Downregulation of miR‐93 in CD44+ cancer cells metastasized to liver of breast cancer PDX mouse
Cancer metastasis requires the seeding, migration, and successful colonization of cancer cells at distant organs; metastatic CSCs with both CSC properties and migrating abilities drive cancer metastasis. 17 To characterize the breast CSCs that are involved in metastatic progression, we analyzed human breast cancer PDX mouse generated by orthotopic xenotransplantation of estrogen receptor(+)/progesterone receptor(+)/HER2(−) human breast cancer tissues. 27 This PDX mouse is characterized by the formation of micrometastases in the liver when the diameter of tumors at the primary site reached approximately 2 cm. To identify miRNAs involved in the metastatic progression of CSCs, the CD44+ human breast cancer cells metastasized to the liver (“metastasized CSCs”) and those in the primary site (“primary CSCs”) were isolated by FACS and purified in parallel from the breast cancer PDX mice (Figure S1). The comparison of the expression profile of 754 miRNAs between the metastasized CSCs and primary CSCs revealed that the expression levels of the miRNAs within the paralogue clusters (miR‐106b‐25, miR‐17‐92, and miR‐106a‐363) were much lower (mostly undetectable) in the metastasized CSCs than in the primary CSCs (Figure 1A,B). These paralogue miRNA clusters are formed by 4 groups of miRNAs with essentially the same seed sequence (ie, a functional sequence for mRNA targeting) (Figure 1A). Given the observed expression patterns, we speculated that these miRNAs function as the suppressor of CD44+ PDX cell metastasis. In this study, we focused on miR‐93 that shares the seed sequence with miR‐106a/b, miR‐20a/b and miR‐17‐5p in the paralogue miRNA clusters, because it was among the miRNAs that were detectable in CD44+ primary CSCs and downregulated (or undetectable) in both CD44− cancer cells at the primary site and CD44+ metastasized CSCs (Figure 1B). Indeed, comparison of the miR‐93 expression levels between the metastasized CSCs and primary CSCs further confirmed the significant downregulation of miR‐93 in the CD44+ cancer cells metastasized to the liver (Figure 1C).
FIGURE 1.
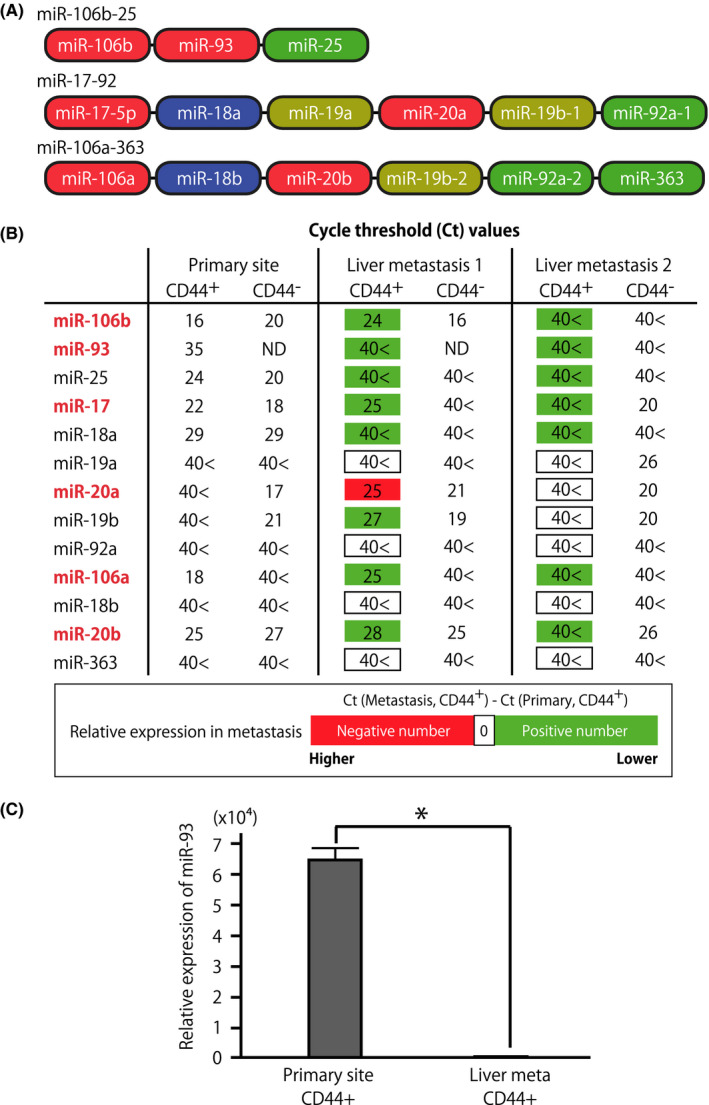
Suppression of microRNA (miR)‐106b‐25 cluster miRNAs in metastatic cancer stem cells in the liver. A, Schematic representation of 3 polycistronic, paralogue clusters (miR‐106b‐25, miR‐17‐92, and miR‐106a‐363). miRNAs were color‐coded based on the similarity of their seed sequences. B, Expression levels of miR‐17‐92, miR‐106a‐363, and miR‐106b‐25 miRNA cluster family miRNAs in the CD44+ and CD44− tumor cells isolated from breast cancer patient‐derived tumor xenograft (PDX) mice. Cancer cells were sorted from the dissociated PDX tumors and the liver of the same PDX mice. Numbers indicate the cycle threshold (Ct) value of semiquantitative PCR, in which lower Ct values indicate higher expression level of the target miRNA. The Ct values were highlighted green or red when the expression level of miRNA in the CD44+ metastasized cells was lower or higher, respectively, than those in CD44+ cancer cells at the primary site. Expression levels of miRNAs were normalized to that of RNU48 snRNA. 40<, undetectable even after 40 cycles of amplification; ND, not determined. C, Downregulation of miR‐93 in CD44+ cancer cells metastasized to the liver. CD44+ cancer cells at the primary site and metastasized to the liver were sorted in parallel from human breast cancer PDX mouse. n = 3, *P < .001
3.2. Inhibition of invasion abilities by miR‐93
Because miR‐93 was downregulated in the CD44+ PDX cells successfully metastasized to the liver, first we evaluated the effect of miR‐93 on cell migration and invasion capacities of breast cancer cells. The results of both wound healing and Boyden chamber‐based transwell cell migration assays showed that overexpression or downregulation of miR‐93 in breast cancer MDA‐MB‐231 cells did not affect their migration abilities (Figures [Link], [Link]). In contrast, in transwell cell invasion assays in which the upper surface of transwell inserts was precoated with Matrigel, overexpression of miR‐93 using miR‐93 mimic significantly reduced the number of hormone‐receptor‐negative (MDA‐MB‐231) and ‐positive (T‐47D) breast cancer cells invaded by 36% and 65%, respectively (Figures 2A and S2); downregulation of miR‐93 using miR‐93 inhibitor in MDA‐MB‐231 and T‐47D cells significantly increased the number of invaded cells by 64% and 76%, respectively (Figures 2B and S2). These results suggest that although its effect on cell migration abilities was not evident, miR‐93 clearly functions as a suppressor of invasion abilities in breast cancer cells.
FIGURE 2.
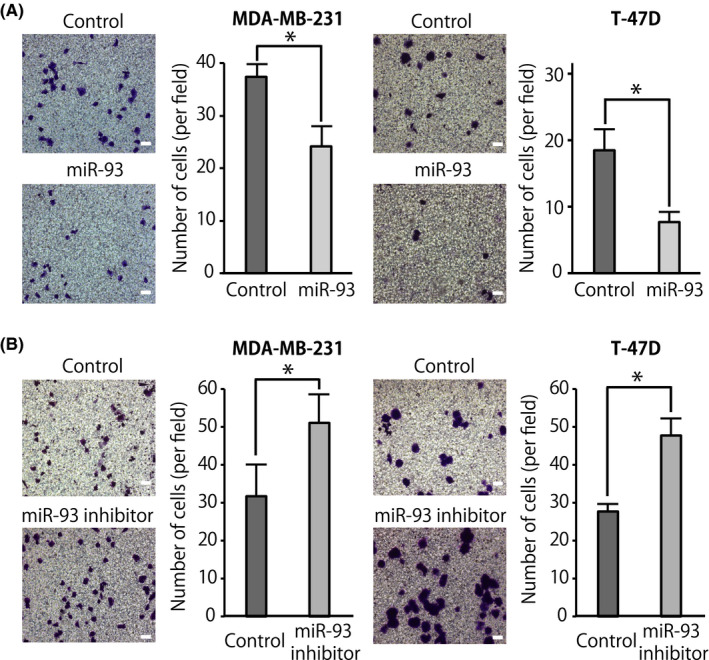
MicroRNA (miR)‐93 suppresses invasion of breast cancer cells. Boyden chamber transwell cell invasion assay was undertaken using breast cancer cells (MDA‐MB‐231 and T‐47D) transfected with miR‐93 mimic or negative control (50 nmol/L) (A) or with miR‐93 inhibitor or negative control (50 nmol/L) (B). Cell invasion was measured after 24 h. Representative micrographs and number of invaded cells in 5 random fields per membrane are shown. Data are presented as mean ± SD. n ≥ 3, *P < .05. Scale bar = 100 μm
3.3. Inhibition of organoid‐forming capacity by miR‐93
To understand whether miR‐93 had a direct mechanistic role in suppressing CSC properties during metastasis, we tested whether miR‐93 was able to affect the 3D organoid‐forming capacity of breast cancer cells. Organoid culture is a 3D culture method that enables ex vivo analysis of stem cell behavior and differentiation. 25 Infection of MDA‐MB‐231 cells with a lentivirus encoding for miR‐93 significantly reduced their capacity to grow as 3D organoids (Figure 3A, upper panels). Consistent with this observation, infection of hormone‐receptor‐positive breast cancer MCF7 cells with a lentivirus encoding for anti‐miR‐93 significantly increased their capacity to grow as 3D organoids (Figure 3A, lower panels). Analyses of the gene expression profiles of organoids showed that forced expression of miR‐93 caused a reduction of the expression levels of stem cell‐related genes and EMT‐related genes, such as CD44, BMI1, and SNAI2 (Figure 3B).
FIGURE 3.
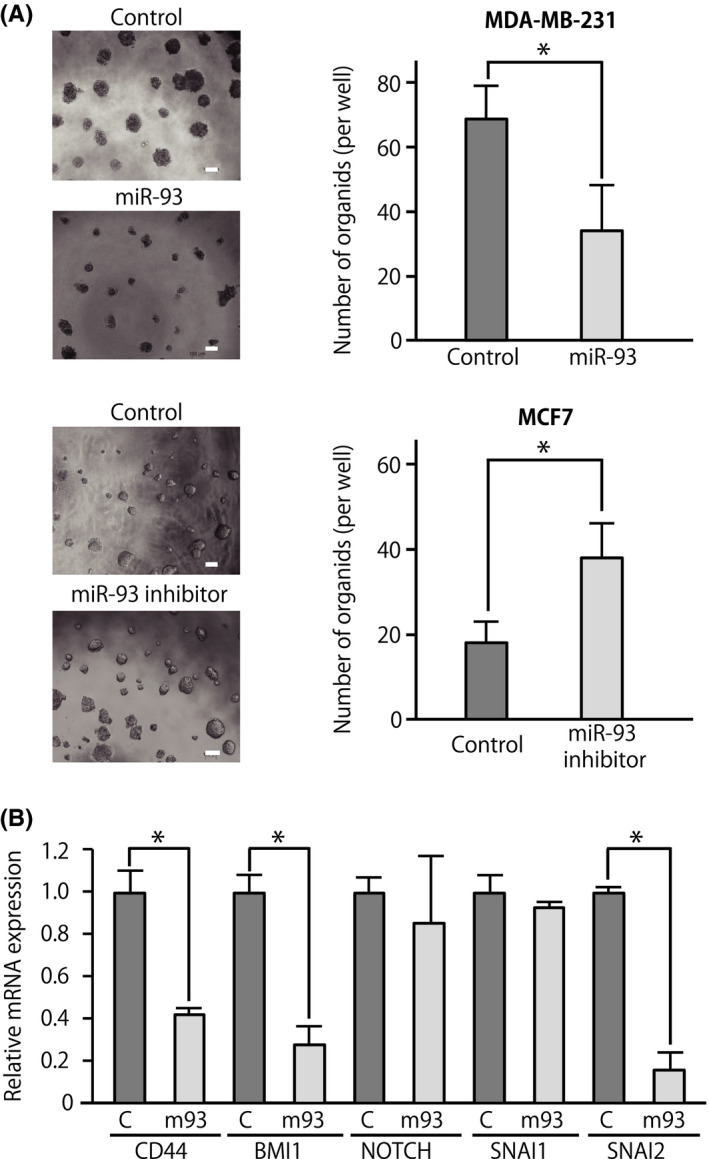
MicroRNA (miR)‐93 suppresses 3D organoid formation capacities. A, 3D organoid‐forming capacity of breast cancer cells (MDA‐MB‐231 and MCF‐7). Cells were infected with a lentivirus vector driving constitutive expression of miR‐93 or its inhibitor. n = 3, *P < .05. Scale bar = 100 μm. B, Expression levels of genes associated with cancer stem cell properties and epithelial‐mesenchymal transition in breast cancer cells forming organoids. C, cells of control organoid; m93, cells of miR‐93‐expressing organoid. Data are presented as mean ± SD. n = 3, *P < .05
3.4. MicroRNA‐93 targets WASF3
Because the results of in vitro analyses suggested that invasion abilities and CSC properties were suppressed by the overexpression of miR‐93, we used the TargetScan 6.2 algorithm (http://www.targetscan.org) to search for putative miR‐93 targets that play important roles in the regulation of CSC properties and cell invasion. The search identified the WASF3 (WAVE3) gene as one of the promising candidates. 28 , 29 Importantly, WASF3 is an actin cytoskeleton remodeling protein, which often functions as an oncogene, and is established as a major driver of tumor progression, invasion (Figure S5 and Table S2), and metastasis of breast cancers, 9 , 29 and is a marker associated with worse patient prognosis. 29 , 30 In addition, WASF3 promotes tumor sphere formation and the expression of CSC‐related genes. 28 Within its 3′‐UTR, WASF3 carries 2 predicted 8‐mer miR‐93 target sites (2728‐2735 and 3889‐3896 of the NM_006646.6): the former is poorly conserved across mammalian species, and the latter is highly conserved across mammalian species (Figure 4A, 8mer in lower boxes). Indeed, miR‐93 overexpression suppressed the luciferase activity driven by the Luc‐WASF3 construct with a highly conserved 8‐mer target sequence (pGL3‐WASF3‐2), but not the one with poorly conserved 8‐mer sequence (pGL3‐WASF3‐1), and this suppressive effect was completely abrogated by the introduction of mutations restricted to the target sequence itself (Figure 4A,B). Moreover, forced expression of miR‐93 in MDA‐MB‐231 cells whose expression level of miR‐93 is relatively low 31 significantly reduced WASF3 protein level, whereas inhibition of miR‐93 in MCF7 cells, which highly express miR‐93, 31 significantly increased its level (Figures 4C and S2). To prove the functional relevance of the WASF3 regulation by miR‐93 in breast cancers, we investigated the invasion abilities of breast cancers using miR‐93 mimic and WASF3‐expression plasmid (Figure 5A). Coexpression of WASF3 abrogated the capacity of miR‐93 to suppress the invasion of breast cancer cells in vitro (Figure 5B). To clarify the functional relevance of the miR‐93‐WASF3 interaction in vivo, we analyzed the breast cancer PDX mice in which the expression level of miR‐93 was significantly downregulated in CD44+ breast cancer cells metastasized to the liver (metastasized CSC) compared to those in the primary site (primary CSC) (Figure 1C). As expected, the expression level of WASF3 mRNA was significantly upregulated in metastasized CSCs compared to primary CSCs (Figure S6).
FIGURE 4.
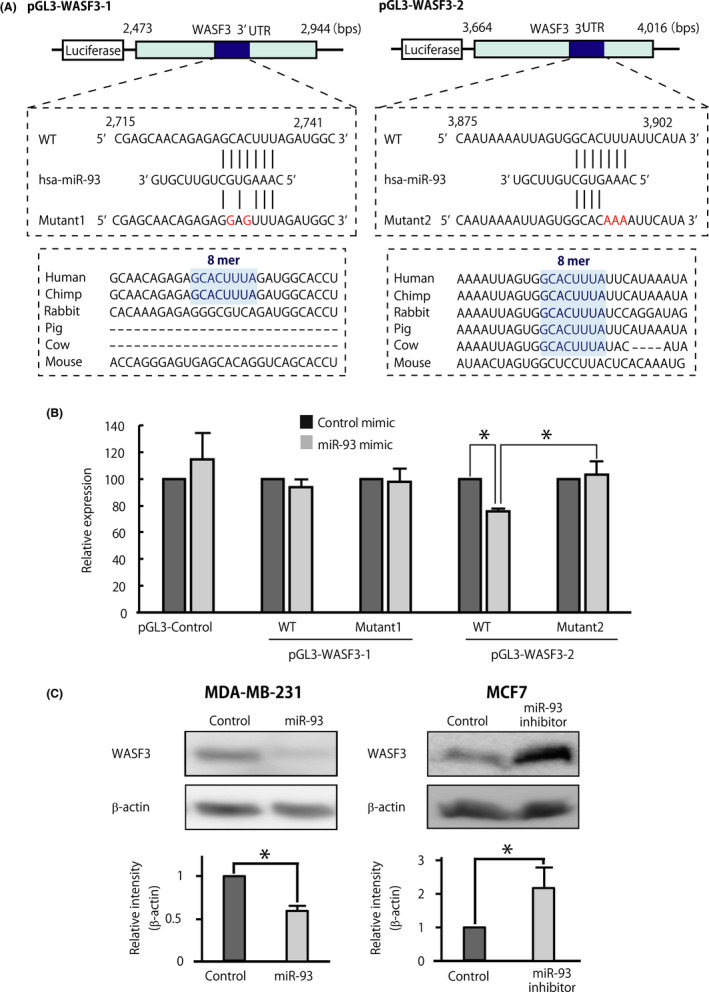
MicroRNA (miR)‐93 directly targets WASF3. A, Schematic representation of the predicted hsa‐miR‐93(‐5p) target recognition sequences within the 3′‐UTR of the WASF3 mRNA, and of the mutations introduced to functionally disable them. Numbers correspond to nucleotide positions in WASF3 sequence (GenBank: NM_006646.6). Lower panels show the conservation levels of nucleotide sequences of the miR‐93 putative target site (8‐mer, highlighted in blue) across multiple mammalian species. B, Luciferase activity of pGL3 constructs encoding the WT or mutant version of the WASF3 3′‐UTR by miR‐93. Luciferase activities were measured after 48 h and normalized by Renilla luciferase activities. Data are presented as mean ± SD. n = 3, *P < .05. C, Endogenous WASF3 protein levels after transfection with miR‐93 mimics or inhibitor (50 nmol/L). Expression of β‐actin was used as a control. Data are presented as mean ± SD. n = 3, *P < .05
FIGURE 5.

Forced expression of WASF3 abrogated microRNA (miR)‐93‐induced inhibition of invasion. A, Expression levels of WASF3 in the MDA‐MB‐231 cells overexpressing control or WASF3 expression plasmid. β‐Actin was used as a loading control. *P < .05. B, Forced expression of WASF3 abrogated the ability of miR‐93 to suppress cell invasion of breast cancer cells (MDA‐MB‐231 and T‐47D) in Boyden chamber transwell cell invasion assay. Cell invasion was measured after 24 h. Representative micrographs and number of migrated cells in 5 random fields per membrane are shown. Data are presented as mean ± SD. n > = 3, *P < .05. Scale bar = 100 μm
3.5. MicroRNA‐93 suppresses the metastatic capacity of breast cancer cells in vivo
Because miR‐93 suppressed the invasion and 3D organoid‐forming capacities and reduced the expression levels of genes associated with stem cell properties and EMT in vitro, we tested whether miR‐93 was able to suppress metastasis of breast cancer cells in immunodeficient NSG mice. Metastatic abilities of breast cancer cells were evaluated using a hepatic metastasis model by a splenic injection of tumor cells using MDA‐MB‐231 cells overexpressing miR‐93 or control MDA‐MB‐231 cells. The results showed that overexpression of miR‐93 in MDA‐MB‐231 cells caused a statistically significant reduction of their in vivo metastatic capacity (Figure 6A,B). Finally, we undertook a competitive transplantation assay to further confirm that expression of miR‐93 affected metastatic capacities of breast cancer cells in vivo. When MDA‐MB‐231‐ZsGreen cells overexpressing miR‐93 (miR‐93 ZsGreen) and MDA‐MB‐231 mCherry competitor (mixed 1:1) were transplanted into the spleen of immunodeficient NSG mice, the percentage of MDA‐MB‐231 mCherry competitor cells was significantly higher than that of miR‐93 ZsGreen cells among the cancer cells successfully colonized to the liver (Figure 6A,C). These results suggest that breast cancer cells expressing miR‐93 had a significant competitive disadvantage in the liver metastasis.
FIGURE 6.
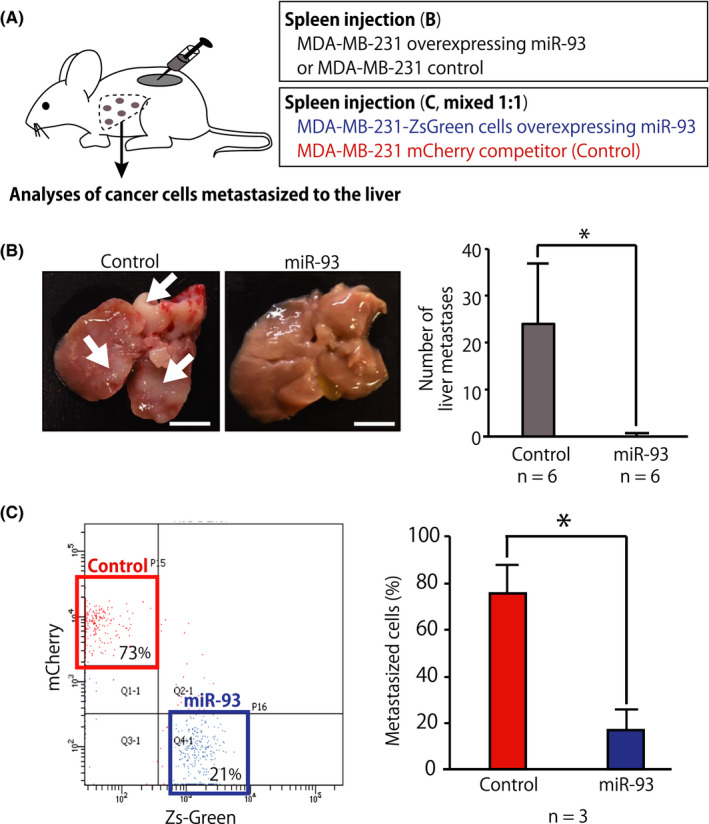
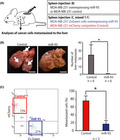
MicroRNA (miR)‐93 suppressed liver metastasis in vivo. A, Schematic representation of the splenic injection of tumor cells and competitive transplantation assays. For the competitive transplantation assay, MDA‐MB‐231‐ZsGreen cells overexpressing miR‐93 (miR‐93 ZsGreen) and MDA‐MB‐231 mCherry competitor (mixed 1:1) were transplanted into the spleen of immunodeficient NSG mice. B, Gross examination of the development of metastases in the liver 21 d after intrasplenic injection of MDA‐MB‐231 cells stably infected with miR‐93‐expression or control lentivirus. Number of tumors on the surfaces of the liver was counted. Arrows indicate representative liver metastases. Data are presented as mean ± SD. n = 6, *P < .05. Scale bar = 10 mm. C, Donor chimerism of the cancer cells metastasized to the liver was evaluated by FACS. The liver was perfused, dissociated, and analyzed 3 wk after transplantation. Percentage of donor chimerism of the cancer cells metastasized in the liver after competitive transplantation assay. Data are presented as mean ± SD. n = 3, *P < .05
4. DISCUSSION
Metastasis is multistep process during which adequate epigenetic changes are required to confer cancer cells the abilities to survive, invade, and successfully colonize to the target organ. 32 Cancer cells with stem cell properties have been proposed to be a driving force of metastasis because they have higher probability to go through these metastatic processes. 33 In this study, we used the breast cancer PDX model to analyze the molecular characteristics of CSC marker CD44+ cancer cells that spontaneously and successfully colonized to the liver (Figure 1). 27 Because tumors of PDX mice are more likely to recapitulate the tumor cell heterogeneity of patient tumor that contains a CSC population, analyses of PDX mouse models enable us to analyze the metastasis of cancer cells with CSC properties. Indeed, a previous report has shown that metastases are initiated by cancer cells with CSC properties in the breast cancer PDX mouse model. 17
The miR‐17‐92 cluster is a highly conserved polycistronic miRNA cluster and has two paralogue gene clusters named miR‐106a‐363 and miR‐106b‐25. The miR‐17‐92 cluster is activated by the Myc proto‐oncogene family and signaling pathways such as Notch and Sonic Hedgehog. The miR‐106b‐25 cluster, which resides in the 13th intron of the MCM7 gene on chromosome 7, is highly conserved across vertebrates. These polycistronic miRNA clusters are highly expressed in a wide range of tumor cells and cancers, such as lung, breast, pancreas, prostate, and thyroid cancer, as well as lymphomas. 34 Therefore, the majority of the previous studies aimed at studying their oncogenic ability to promote tumor cell proliferation, invasion, and metastases, and much fewer studies delineated antitumor properties of these clusters. 35
Our results showed that downregulation of miR‐93, a member of the miR‐106b‐25 cluster, is associated with a part of the multistep metastasis processes, namely invasion and maintenance of CSC properties, but not cell migration. Previous reports have shown that miR‐93 is able to both enhance and inhibit metastasis depending on the cellular context, at least partly because miR‐93 targets multiple target genes that either promote or suppress metastasis. 36 MicroRNA‐93 promotes cell invasion and metastasis by targeting PTEN, NEDD4L, and TGFBR2, which are the regulators of cell migration, transforming growth factor‐β signaling, and EMT. 37 , 38 In contrast, miR‐93 inhibits EMT in breast cancer cells by targeting MKL1, STAT3, and WNK lysine deficient protein kinase 1 (WNK1). 39 , 40 In this study, we showed that miR‐93 suppressed invasion abilities of MDA‐MB‐231 cells in a WASF3 suppression‐dependent manner (Figures 2 and 5). However, its effect on cell migration ability, which was less associated with protease‐induced digestion of matrices, was not evident with transwell migration and would healing assays (Figures S3 and S4). Because suppression of WASF3 enhances the expression level of MMPs, 41 , 42 , 43 we speculate that the effects of miR‐93 on invasion of breast cancer cells became more evident than its effects on migration in our in vitro experimental settings.
In addition to being a specific effector downstream of the Rac GTPases in actin polymerization, 44 , 45 , 46 WASF3 is involved in the regulation of stem cell properties; although the molecular mechanisms involved in the regulation of stem cell properties are not fully elucidated. For example, knockdown of WASF proteins impairs bone marrow repopulation by hematopoietic stem cells, 47 decreases mammosphere formation capacity after serial passaging, 28 and attenuates the expression of CSC maintenance genes in breast cancer cells by suppressing the translocation of Y‐box‐binding protein‐1. 28 Therefore, it is possible that WASF3 is one of the targets of miR‐93 for the suppression of the CSC properties of breast cancer cells (Figure 3). Taken together, our results indicate that downregulation of miR‐93 is one of the factors for an enhancement of both cell invasion capacity and CSC properties, which are required for successful colonization of CSCs to distant organs.
CONFLICT OF INTEREST
YS is listed as a co‐inventor on a patent application that describes the use of miRNAs as biomarkers for the identification and therapeutic targeting of cancer stem cells (US‐20110021607). NS and YK declare no competing interests.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Table S1
Table S2
Appendix S1
ACKNOWLEDGMENTS
We thank Drs. Miki Nishio, Jyunji Otani, Junko Mukohyama, Tomohiko Maehama and Tadaomi Takenawa (Kobe University, Japan), Taichi Isobe (Stanford University, USA), Masao Maeda and Takanori Hayashi (Fujita Health University, Japan), and Tatsunori Nishimura and Noriko Gotoh (Kanazawa University, Japan) for their superior support and discussions, Seishi Kono and Shintaro Takao (Kobe University) for collection of clinical specimens, and Hironobu Minami (Kobe University) for his superior mentorship. This work was supported by grants from the Japan Society for the Promotion of Science (JSPS KAKENHI) (15K14381, 18K07231, and Japan‐Belgium Research Cooperative Program to YS), the Princess Takamatsu Cancer Research Fund (to YS), the Fujita Health University (to YS), and the Cancer Research Institute of Kanazawa University (to YS).
Shibuya N, Kakeji Y, Shimono Y. MicroRNA‐93 targets WASF3 and functions as a metastasis suppressor in breast cancer. Cancer Sci. 2020;111:2093–2103. 10.1111/cas.14423
REFERENCES
- 1. O'Shaughnessy J. Extending survival with chemotherapy in metastatic breast cancer. Oncologist. 2005;10(Suppl 3):20‐29. [DOI] [PubMed] [Google Scholar]
- 2. Celia‐Terrassa T, Kang Y. Distinctive properties of metastasis‐initiating cells. Genes Dev. 2016;30:892‐908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim J, Yao F, Xiao Z, Sun Y, Ma L. MicroRNAs and metastasis: small RNAs play big roles. Cancer Metastasis Rev. 2018;37:5‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma L, Teruya‐Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA‐10b in breast cancer. Nature. 2007;449:682‐688. [DOI] [PubMed] [Google Scholar]
- 5. Kim J, Siverly AN, Chen D, et al. Ablation of miR‐10b suppresses oncogene‐induced mammary tumorigenesis and metastasis and reactivates tumor‐suppressive pathways. Cancer Res. 2016;76:6424‐6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ma LI, Young J, Prabhala H, et al. miR‐9, a MYC/MYCN‐activated microRNA, regulates E‐cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen D, Sun Y, Wei Y, et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo‐YAP pathway and a prognostic marker. Nat Med. 2012;18:1511‐1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang L, Zhang S, Yao J, et al. Microenvironment‐induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527:100‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teng Y, Qin H, Bahassan A, Bendzunas NG, Kennedy EJ, Cowell JK. The WASF3‐NCKAP1‐CYFIP1 complex is essential for breast cancer metastasis. Cancer Res. 2016;76:5133‐5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qin H, Lu S, Thangaraju M, Cowell JK. Wasf3 deficiency reveals involvement in metastasis in a mouse model of breast cancer. Am J Pathol. 2019;189:2450‐2458. [DOI] [PubMed] [Google Scholar]
- 11. Teng Y, Mei Y, Hawthorn L, Cowell JK. WASF3 regulates miR‐200 inactivation by ZEB1 through suppression of KISS1 leading to increased invasiveness in breast cancer cells. Oncogene. 2014;33:203‐211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bracken CP, Gregory PA, Kolesnikoff N, et al. A double‐negative feedback loop between ZEB1‐SIP1 and the microRNA‐200 family regulates epithelial‐mesenchymal transition. Cancer Res. 2008;68:7846‐7854. [DOI] [PubMed] [Google Scholar]
- 13. Wellner U, Schubert J, Burk UC, et al. The EMT‐activator ZEB1 promotes tumorigenicity by repressing stemness‐inhibiting microRNAs. Nat Cell Biol. 2009;11:1487‐1495. [DOI] [PubMed] [Google Scholar]
- 14. Cui YX, Bradbury R, Flamini V, Wu B, Jordan N, Jiang WG. MicroRNA‐7 suppresses the homing and migration potential of human endothelial cells to highly metastatic human breast cancer cells. Br J Cancer. 2017;117:89‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shen L, Wang P, Yang J, Li X. MicroRNA‐217 regulates WASF3 expression and suppresses tumor growth and metastasis in osteosarcoma. PLoS ONE. 2014;9:e109138. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Shimono Y, Mukohyama J, Nakamura S, Minami H. MicroRNA regulation of human breast cancer stem cells. J Clin Med. 2015;5:E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawson DA, Bhakta NR, Kessenbrock K, et al. Single‐cell analysis reveals a stem‐cell program in human metastatic breast cancer cells. Nature. 2015;526:131‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shimono Y, Zabala M, Cho RW, et al. Downregulation of miRNA‐200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592‐603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Al‐Hajj M, Wicha MS, Benito‐Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983‐3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Isobe T, Hisamori S, Hogan DJ, et al. miR‐142 regulates the tumorigenicity of human breast cancer stem cells through the canonical WNT signaling pathway. Elife. 2014;3:e01977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu F, Yao H, Zhu P, et al. let‐7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109‐1123. [DOI] [PubMed] [Google Scholar]
- 22. Mukohyama J, Isobe T, Hu Q, et al. miR‐221 targets QKI to enhance the tumorigenic capacity of human colorectal cancer stem cells. Cancer Res. 2019;79:5151‐5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goto H, Shimono Y, Funakoshi Y, et al. Adipose‐derived stem cells enhance human breast cancer growth and cancer stem cell‐like properties through adipsin. Oncogene. 2019;38:767‐779. [DOI] [PubMed] [Google Scholar]
- 24. Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241‐245. [DOI] [PubMed] [Google Scholar]
- 25. Shimono Y, Mukohyama J, Isobe T, Johnston DM, Dalerba P, Suzuki A. Organoid culture of human cancer stem cells. Methods Mol Biol. 2019;1576:23‐31. [DOI] [PubMed] [Google Scholar]
- 26. Soares KC, Foley K, Olino K, et al. A preclinical murine model of hepatic metastases. J Vis Exp. 2014;91:51677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nobutani K, Shimono Y, Mizutani K, et al. Downregulation of CXCR4 in metastasized breast cancer cells and implication in their dormancy. PLoS ONE. 2015;10:e0130032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bledzka K, Schiemann B, Schiemann WP, Fox P, Plow EF, Sossey‐Alaoui K. The WAVE3‐YB1 interaction regulates cancer stem cells activity in breast cancer. Oncotarget. 2017;8:104072‐104089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kulkarni S, Augoff K, Rivera L, et al. Increased expression levels of WAVE3 are associated with the progression and metastasis of triple negative breast cancer. PLoS ONE. 2012;7:e42895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Teng Y, Pi W, Wang Y, Cowell JK. WASF3 provides the conduit to facilitate invasion and metastasis in breast cancer cells through HER2/HER3 signaling. Oncogene. 2016;35:4633‐4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Riaz M, van Jaarsveld MTM, Hollestelle A, et al. miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation‐specific miRNAs. Breast Cancer Res. 2013;15:R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liao WT, Ye YP, Deng YJ, Bian XW, Ding YQ. Metastatic cancer stem cells: from the concept to therapeutics. Am J Stem Cells. 2014;3:46‐62. [PMC free article] [PubMed] [Google Scholar]
- 34. Xiang J, Wu J. Feud or friend? The role of the miR‐17‐92 cluster in tumorigenesis. Curr Genomics. 2010;11:129‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fang L‐L, Wang X‐H, Sun B‐F, et al. Expression, regulation and mechanism of action of the miR‐17‐92 cluster in tumor cells (Review). Int J Mol Med. 2017;40:1624‐1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mehlich D, Garbicz F, Wlodarski PK. The emerging roles of the polycistronic miR‐106b approximately 25 cluster in cancer – a comprehensive review. Biomed Pharmacother. 2018;107:1183‐1195. [DOI] [PubMed] [Google Scholar]
- 37. Li N, Miao Y, Shan Y, et al. MiR‐106b and miR‐93 regulate cell progression by suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell Death Dis. 2017;8:e2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qu MH, Han C, Srivastava AK, et al. miR‐93 promotes TGF‐beta‐induced epithelial‐to‐mesenchymal transition through downregulation of NEDD4L in lung cancer cells. Tumour Biol. 2016;37:5645‐5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiang Y, Liao X‐H, Yu C‐X, et al. MiR‐93‐5p inhibits the EMT of breast cancer cells via targeting MKL‐1 and STAT3. Exp Cell Res. 2017;357:135‐144. [DOI] [PubMed] [Google Scholar]
- 40. Shyamasundar S, Lim JP, Bay BH. miR‐93 inhibits the invasive potential of triple‐negative breast cancer cells in vitro via protein kinase WNK1. Int J Oncol. 2016;49:2629‐2636. [DOI] [PubMed] [Google Scholar]
- 41. Teng Y, Liu M, Cowell JK. Functional interrelationship between the WASF3 and KISS1 metastasis‐associated genes in breast cancer cells. Int J Cancer. 2011;129:2825‐2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Teng Y, Ren MQ, Cheney R, Sharma S, Cowell JK. Inactivation of the WASF3 gene in prostate cancer cells leads to suppression of tumorigenicity and metastases. Br J Cancer. 2010;103:1066‐1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Teng Y, Bahassan A, Dong D, et al. Targeting the WASF3‐CYFIP1 complex using stapled peptides suppresses cancer cell invasion. Cancer Res. 2016;76:965‐973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14:7‐12. [DOI] [PubMed] [Google Scholar]
- 45. Kurisu S, Takenawa T. WASP and WAVE family proteins: friends or foes in cancer invasion? Cancer Sci. 2010;101:2093‐2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mendoza MC. Phosphoregulation of the WAVE regulatory complex and signal integration. Semin Cell Dev Biol. 2013;24:272‐279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ogaeri T, Eto K, Otsu M, Ema H, Nakauchi H. The actin polymerization regulator WAVE2 is required for early bone marrow repopulation by hematopoietic stem cells. Stem Cells. 2009;27:1120‐1129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Table S1
Table S2
Appendix S1


