We present here evidence of the existence of the mandelate pathway in yeast for the synthesis of benzenoids. The link between phenylpyruvate- and 4-hydroxyphenlypyruvate-derived compounds with the corresponding synthesis of benzaldehydes through benzoylformate decarboxylation is demonstrated. Hanseniaspora vineae was used in these studies because of its capacity to produce benzenoid derivatives at a level 2 orders of magnitude higher than that produced by Saccharomyces. Contrary to what was hypothesized, neither β-oxidation derivatives nor 4-coumaric acid is an intermediate in the synthesis of yeast benzenoids. Our results might offer an answer to the long-standing question of the origin of 4-hydroxybenzoate for the synthesis of Q10 in humans.
KEYWORDS: 4-hydroxybenzoic acid, ARO10, Hanseniaspora vineae, benzoylformate decarboxylase, benzyl alcohol, coenzyme Q, phenylpyruvate pathway
ABSTRACT
Benzenoid-derived metabolites act as precursors for a wide variety of products involved in essential metabolic roles in eukaryotic cells. They are synthesized in plants and some fungi through the phenylalanine ammonia lyase (PAL) and tyrosine ammonia lyase (TAL) pathways. Ascomycete yeasts and animals both lack the capacity for PAL/TAL pathways, and metabolic reactions leading to benzenoid synthesis in these organisms have remained incompletely known for decades. Here, we show genomic, transcriptomic, and metabolomic evidence that yeasts use a mandelate pathway to synthesize benzenoids, with some similarities to pathways used by bacteria. We conducted feeding experiments using a synthetic fermentation medium that contained either 13C-phenylalanine or 13C-tyrosine, and, using methylbenzoylphosphonate (MBP) to inhibit benzoylformate decarboxylase, we were able to accumulate intracellular intermediates in the yeast Hanseniaspora vineae. To further confirm this pathway, we tested in separate fermentation experiments three mutants with deletions in the key genes putatively proposed to form benzenoids (Saccharomyces cerevisiae aro10Δ, dld1Δ, and dld2Δ strains). Our results elucidate the mechanism of benzenoid synthesis in yeast through phenylpyruvate linked with the mandelate pathway to produce benzyl alcohol and 4-hydroxybenzaldehyde from the aromatic amino acids phenylalanine and tyrosine, as well as sugars. These results provide an explanation for the origin of the benzoquinone ring, 4-hydroxybenzoate, and suggest that Aro10p has benzoylformate and 4-hydroxybenzoylformate decarboxylase functions in yeast.
IMPORTANCE We present here evidence of the existence of the mandelate pathway in yeast for the synthesis of benzenoids. The link between phenylpyruvate- and 4-hydroxyphenlypyruvate-derived compounds with the corresponding synthesis of benzaldehydes through benzoylformate decarboxylation is demonstrated. Hanseniaspora vineae was used in these studies because of its capacity to produce benzenoid derivatives at a level 2 orders of magnitude higher than that produced by Saccharomyces. Contrary to what was hypothesized, neither β-oxidation derivatives nor 4-coumaric acid is an intermediate in the synthesis of yeast benzenoids. Our results might offer an answer to the long-standing question of the origin of 4-hydroxybenzoate for the synthesis of Q10 in humans.
INTRODUCTION
Benzenoid-derived metabolites are of particular importance in plants, as they serve as precursors to a wide variety of compounds involved in essential roles, such as hormone production, defense, and molecules involved in plant-insect communication (1). Benzyl alcohol is a compound widely used in the cosmetic, pharmaceutical, and fragrance industries (2) and may also play roles in the cell-cell interactions of fungi (3, 4) and supply of hydrogen peroxide for lignin biodegradation (5). In both human and yeast cells, the 4-hydroxybenzoic acid (4-HB) aromatic ring precursor plays an important role in the synthesis of coenzyme Q, which is an essential component in the respiratory electron transport chain of the mitochondria (6, 7) (Q6 in Saccharomyces [8, 9], Q9 in mice, and Q10 in humans [6, 10, 11]). Still, after more than 5 decades, little is known about the metabolic pathways that lead to the formation of volatile benzenoids in mammals and yeast (9, 12, 13). It has been established that benzyl alcohol is formed in plants and some fungi during the synthesis of phenylpropanoids via the phenylalanine ammonia lyase (PAL) pathway (14–16). PAL catalyzes the conversion of phenylalanine (Phe) to trans-cinnamic acid, which is subsequently converted into benzaldehyde and other derived compounds through oxidative and nonoxidative pathways (1). In plants, PAL is involved in the biosynthesis of p-coumarate and kaempferol, both of which are precursors to 4-HB (17, 18), and has been identified in some fungi of both Basidiomycota and Ascomycota, including Neurospora, Aspergillus, and Botrytis (14); however, this enzyme has not been identified in the Saccharomycotina subphylum. The formation of 4-HB in yeast and mammalian cells suggests that an unknown pathway exists (9). Interestingly, supplementation with 4-HB may have potential for correcting Q10 deficiencies in humans, as 4-HB is more hydrophobic and, thus, has higher bioavailability than coenzyme Q10 itself (9, 19). The initial and final reactions in a putative pathway of benzenoid synthesis in yeast have recently been elucidated, which correspond to the deamination of tyrosine (Tyr) to 4-hydroxyphenyl pyruvate (involving Aro8p and Aro9p) (20, 21) and the oxidation of 4-hydroxybenzaldehyde to 4-HB by Hfd1p (21), respectively. The latter reaction has also proven to be the function of the human ALDH3A1 gene when expressed in yeast. Thus, the complete metabolic pathway(s) leading to the formation of volatile benzenoids in both yeasts and animals remains unclear (Fig. 1) (10).
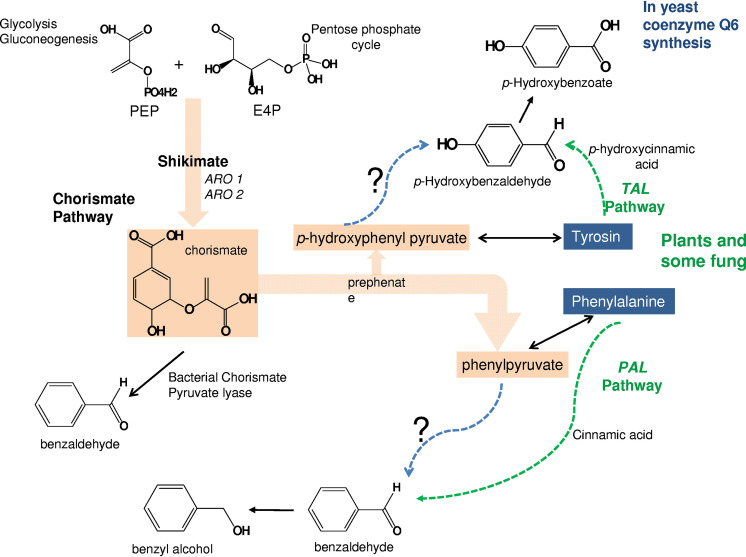
FIG 1.
In plants and some fungi, the phenylalanine ammonia lyase/tyrosine ammonia lyase (PAL/TAL, dotted lines) activities catalyze the conversion of Phe or Tyr into trans-cinnamic acid (28) and trans-hydroxycinnamic acid, respectively, which are subsequently converted into benzyl alcohol or 4-hydroxybenzoic acid and other derived compounds through oxidative and nonoxidative pathways (1, 44). The shikimate and chorismate pathway for the synthesis of aromatic amino acids is shown. The main intermediates are indicated: phenlypyruvate and p-hydroxyphenylpyruvate. The PAL/TAL reactions have not been demonstrated in Saccharomycotina yeasts, bacteria, and animal cells (1, 14, 23). In bacteria, they synthesize 4-HB directly from chorismate (45) or, in the case of Pseudomonas putida, through a mandelate pathway (46). The great production of benzyl-derived compounds by H. vineae through an alternative pathway will contribute to our understanding of the steps that are shown with interrogation symbols on this scheme. PEP, phosphoenolpyruvate; E4P, erythrose 4-phosphate.
The successful metabolic performance (with regard to increased fruity flavors) of Hanseniaspora vineae in wine fermentation (22, 23) has aided the discovery of strains capable of producing benzenoids at a concentration of up to 1,000 μg/liter; this is compared to other yeast species that produce only a few micrograms per liter (20, 24). In fact, when low-assimilable-nitrogen grape musts (under 100 mg N/liter) were used for fermentation, H. vineae has been shown to produce higher concentrations of benzenoids than have been reported in plants (25). Furthermore, in the current study, using a fermentation medium containing even lower levels of ammonia along with increased sources of Phe and Tyr, we were able to precisely measure the extracellular benzenoids produced by Saccharomyces cerevisiae. We conducted experiments using methylbenzoylphosphonate (MBP) to inhibit benzoylformate decarboxylase, the key enzyme in the mandelate pathway of bacteria (26), and we were able to accumulate intracellular intermediates in yeast. While an alternative pathway has previously been hypothesized to exist in plants (1) and some fungi (27), attempts at detailing its mechanism have failed, likely due to technical limitations of the studies (28), as well as the coexistence of the PAL/TAL pathway alternative. Results presented here make possible the exploration of this metabolic pathway in other eukaryotic cells through the study of benzoylformate decarboxylase function.
RESULTS
Enzymatic test for PAL/TAL activity in cell extracts of H. vineae.
To confirm there was no promiscuous activity by PAL or tyrosine ammonia lyase (TAL) in H. vineae cells, we conducted an enzymatic test by incubating cell extracts with high concentrations of Phe and Tyr. Gas chromatography-mass spectrometry (GC-MS) analysis demonstrated that no cinnamic acids were formed by these H. vineae cell extracts. As a positive control for this procedure, we added extra Phe and Tyr to Vitis vinifera grape extracts in vitro and were able to consistently demonstrate the formation of coumarate.
13C-Phe and 13C-Tyr feeding experiments.
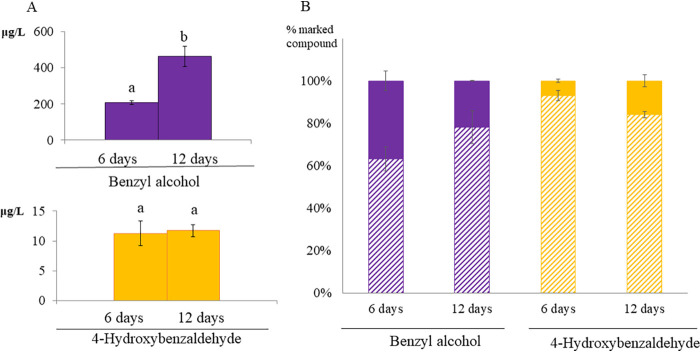
Fermentations of H. vineae performed with 13C-Phe or 13C-Tyr in the synthetic media allowed the detection of extracellular benzyl alcohol and 4-hydroxybenzaldehyde, respectively, which were isotopically marked from day 6. Concentrations of these compounds continued to increase until day 12 of fermentation (Fig. 2A); however, a decrease in the percentage of labeled 13C-benzyl alcohol (Fig. 2B) clearly indicated these compounds were also being synthesized from sugars via the shikimate pathway from the middle to the end of the fermentation. In contrast, an increase in the percent labeled 4-hydroxybenzaldehyde is seen at day 12, but the concentration of this compound is 60-fold less than that for benzyl alcohol (Fig. 2A).
FIG 2.
Feeding experiments using 13C-Phe and 13C-Tyr. (A) Extracellular accumulation of benzenoids in H. vineae increased during fermentation until all the sugars were consumed after 12 days. The Tyr-derived compound pathway accumulated significantly lower concentrations (yellow bars) than the Phe-derived compounds (violet bars) (20). (B) The percentage of marked (solid violet) and unmarked (striped violet) extracellular benzyl alcohol produced in experiments with the addition of 13C-Phe, and marked (solid yellow) and unmarked (striped yellow) extracellular 4-hydrozybenzaldehyde produced in experiments with the addition of 13C-Tyr, following fermentation for 6 or 12 days. Values represent means ± standard deviations (SD), n = 3 independent experiments.
MBP inhibition of putative benzoylformate decarboxylases in H. vineae (HvARO10).
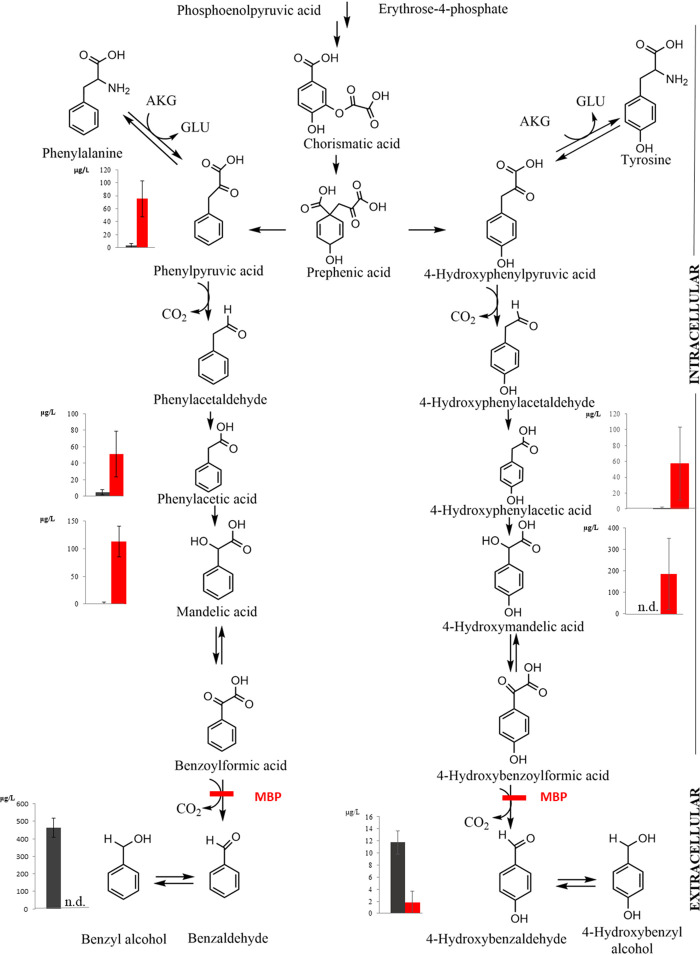
In search of the reactions responsible for the formation of benzaldehyde and putative involvement of the HvARO10 gene, we synthesized a compound analogous to benzoylformic acid, namely, MBP, which has previously been shown to block the benzoylformate decarboxylase enzyme of Pseudomonas putida (26). MBP was synthesized following the method previously described by Brandt et al. (29). We reasoned that extracellular and intracellular metabolic analyses on fermentations including MBP would provide evidence for or against the synthesis of benzenoid intermediates through the mandelate pathway. Thiamine and magnesium were included in this medium, as detailed in Materials and Methods. Aro10p belongs to the group of thiamine diphosphate-dependent decarboxylases, including benzoylformate decarboxylase, and it has been well established that these enzymes require Mg2+ as a cofactor (30). As presented in Fig. 3, we observed the intracellular accumulation of mandelic, phenylacetic, and phenylpyruvic acids, demonstrating that these compounds are intermediates in this benzenoid pathway. Although we did not detect phenylacetaldehyde in our experiments, this compound remains an obvious intermediate in the chemical reactions and may have avoided detection due to its instability under fermentation conditions. Similar intracellular accumulations of 4-hydroxyphenylpyruvate-derived compounds were found, namely, 4-hydroxyphenylacetic and 4-hydroxymandelic acids. Taken together, these results demonstrate that the phenylpyruvate/mandelate pathway is used by H. vineae for the synthesis of benzenoids.
FIG 3.
Phenylpyruvate/mandelate pathway in yeast demonstrating the effect of methyl benzoylphosphonate (MBP), an analogue of naturally occurring benzoylformic acid. Bar graphs represent the intra- and extracellular concentrations of metabolites (following 6 and 12 days of fermentation, respectively) involved in benzyl alcohol and 4-hydroxybenzaldehyde production by H. vineae. The gray bars represent various concentrations in the pathway without addition of the inhibitor, while the red bars represent the metabolite concentrations when MBP was included from the beginning of fermentation. The inhibition of benzoylformic acid decarboxylase by MBP resulted in increased concentrations of most intermediates in the pathway. The two aldehydes (phenylacetaldehyde and 4-hydroxyphenylacetaldehyde) were not detected; however, they were included anyway as putative metabolites in the reactions. AKG, alpha ketoglutarate; GLU, glutamic acid. Values represent means ± SD, n = 3 independent experiments. n.d., not determined.
Deletion mutations in key putative genes of the mandelate pathway validate the effects of MBP in S. cerevisiae.
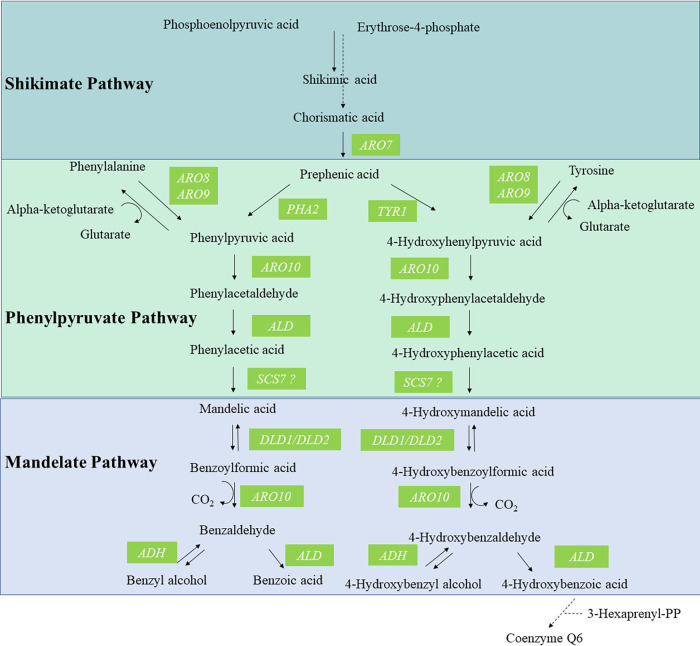
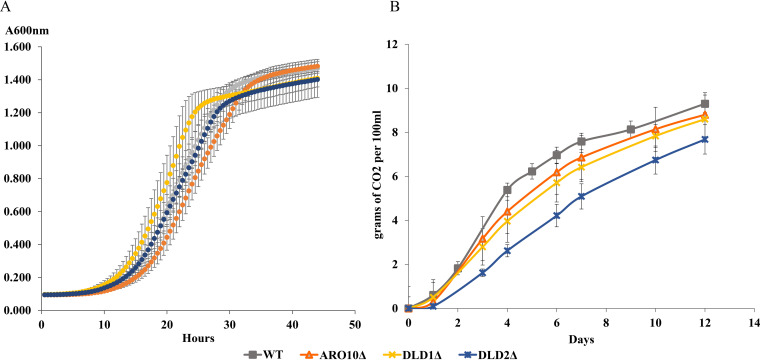
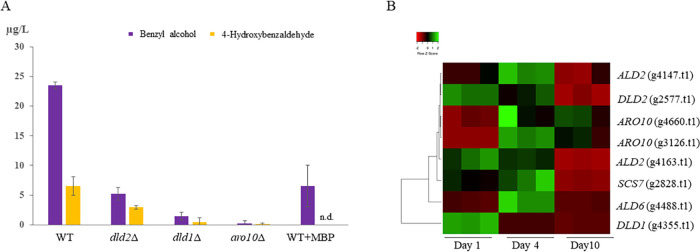
To further confirm the existence of the mandelate pathway, we analyzed three S. cerevisiae deletion mutations in enzymes putatively involved in the pathway according to our genomic studies comparing H. vineae to S. cerevisiae (Table 1 and Fig. 4). Previous reports in yeast have additionally supported these proposed activities of ARO10 (30) and DLD (31, 32). We optimized benzenoid production in a synthetic medium with low assimilable nitrogen content using the wild-type (WT) strain of S. cerevisiae and performed independent fermentations over 12 days with aro10Δ, dld1Δ, and dld2Δ mutants. The fermentation curves, measured as evolved CO2 weight loss, displayed behavior similar between all three mutant strains and to that of the WT (Fig. 5). Fermentation using the mutant strains resulted in no or significantly reduced production of benzyl alcohol and 4-hydroxybenzaldehyde compared to that of the BY4743 WT strain (Fig. 6A). Under our fermentation conditions, we observed 5- and 10-fold decreases in the extracellular levels of benzyl alcohol in the mutant strains dld2Δ and dld1Δ compared with that of the WT, respectively, while the aro10Δ mutant produced no benzenoids. The significantly decreased benzenoid synthesis in the three mutant strains confirms that these enzymes are involved in the mandelate pathway in S. cerevisiae. The addition of MBP at the beginning of fermentation with WT S. cerevisiae resulted in a 4-fold reduction in benzyl alcohol production compared to control fermentations and completely blocked 4-hydroxybenzaldehyde production, validating the inhibitory effect of MBP in S. cerevisiae (Fig. 6A).
TABLE 1.
Main genes of H. vineae compared to the S. cerevisiae genome related to the benzenoid synthetic pathwaya
| Biosynthetic route | Enzymatic activity | Genes identified in H. vineae (% amino acid identity) |
|---|---|---|
| Shikimate pathway | Synthesis of chorismate, phenylalanine, tyrosine | ARO1 (66.79); ARO2 (80.59); ARO3 (77.03); ARO4 (83.51); ARO7 (67.97); PHA2 (41.99); TYR1 |
| Phenylpyruvate and 4-hydroxyphenylpyruvate pathways | Aromatic amino acid transferases | 3×ARO8 (45.51, 59.84, 56.06); 4×ARO9 (42.70, 35.27, 36.08, 36.91) |
| Decarboxylase, aldehyde dehydrogenase, hydrolase | 2×ARO10 (34.10, 30.99); 2×ALD2 (40.55, 44.01); ALD5 (53.45); ALD6 (55.07); SCS7 (66.50) | |
| Mandelate pathway | Dehydrogenase, benzoylformate decarboxylase | DLD1 (53.00); DLD2 (70.00); 2×ARO10 (34.10, 30.99) |
| Benzyl/4-hydroxybenzyl alcohol synthesis | Alcohol dehydrogenases | 2×ADH1 (77.71, 78.74); 2×ADH3 (74.79, 74.80); 4×ADH6 (44.74, 44.47, 44.74, 44.06); SFA1 (68.16) |
| Benzoic and 4-HB acid synthesis from benzaldehyde | Aldehyde dehydrogenases | 2×ALD2 (40.55, 44.01); ALD5 (53.45); ALD6 (55.07) |
Genes present in H. vineae are indicated, and the presence of more than one copy is indicated as 2×, 3×, or 4×. Percentages in parentheses indicate amino acid identity between both strain species. Benzoic acid in plants is synthesized by a benzaldehyde dehydrogenase, BALDH, that is not found in yeast, although the function exists in S. cerevisiae and humans, and some ALD genes may be involved (21, 48–50).
FIG 4.
Proposed reactions in the phenylpyruvate and mandelate pathways in yeast and genes proposed to be involved in the phenylpyruvate/mandelate pathway for synthesis of benzenoids. Interestingly, it has been reported that these enzymes in bacteria might use both phenylpyruvate- and 4-hydroxyphenylpyruvate-derived compounds as substrates (47), and our results also suggest a double function in yeast.
FIG 5.
(A) Growth of S. cerevisiae WT and deletion mutants in fermentation medium measured by absorbance at 600 nm for 48 h. (B) Fermentation kinetics of the S. cerevisiae WT and deletion mutants for 12 h. Bars represent standard deviations. No significant differences were found between strains.
FIG 6.
(A) Benzyl alcohol and 4-hydroxybenzaldehyde production after 12 days of fermentation with either WT S. cerevisiae or a mutant with the deletion of one of three key genes in the pathway (dld2Δ, dld1Δ, and aro10Δ). To validate the existence of benzoylformic acid decarboxylase in S. cerevisiae, the WT strain was treated with the specific inhibitor MBP. Significantly decreased benzenoid formation was observed, mostly due to decreased production of Tyr-derived compounds (4-hydroxybenzaldehyde). Values represent means ± SD, n = 3 independent experiments. H. vineae is known to produce benzenoids at concentrations approximately 100 times greater than those of S. cerevisiae strains in standard grape must fermentations (20). We were able to increase the production of extracellular benzyl alcohol and 4-hydroxybenzaldehyde by S. cerevisiae in a low-yeast-assimilable-nitrogen synthetic medium (YAN; 100 mg N/liter). (B) Transcriptome analysis of eight putative genes of the phenylpyruvate/mandelate pathway in H. vineae (Table 1). The heatmap demonstrates relative gene expression at three stages of the fermentation process (days 1, 4, and 10) and shows consistent expression of all the genes. The two copies of ARO10 may explain the higher activity of benzoylformic acid decarboxylation in this species compared to that in S. cerevisiae.
Expression analysis of key genes relating to the phenylpyruvate/mandelate pathway in H. vineae.
The gene analysis detailed in Fig. 4 and Table 1 was conducted to determine the expression of key genes relating to the phenylpyruvate/mandelate pathway in H. vineae. The transcriptomic profiles of H. vineae were analyzed on three different days of the fermentation process, representing the exponential growth phase (day 1), the end of the exponential phase (day 4), and the end of the stationary phase (day 10). Eight genes predicted to be involved in the phenylpyruvate/mandelate pathway were differentially expressed between at least two of these stages (Fig. 6B). ALD and SCS7, both putatively involved in successive steps of the oxidation of phenylacetaldehyde into mandelic acid (20), demonstrated similar expression patterns, including maximum expression on day 4. It is noteworthy that the ALD2 gene presents two copies with different expression patterns; one of them is highly expressed on day 4 and the other presents high expression on days 1 and 4. In contrast, DLD1 and DLD2 were both highly expressed on day 1, after which their expression levels were reduced throughout the fermentation process. Both copies of ARO10 demonstrated similar expression patterns, with increases in their expression on days 4 and 10, a finding that was in agreement with benzenoid formation.
DISCUSSION
H. vineae is an ancestral species of Saccharomycotina (33) that, compared with S. cerevisiae, has a higher capacity for benzenoid synthesis due to an increased gene copy number of the ARO gene family (23) (Table 1). The fact that H. vineae, an apiculate yeast, produced benzenoid compounds in amounts approximately 2 orders of magnitude larger than that of Saccharomyces suggests its potential use as an ideal, novel eukaryotic model for studying phenylpropanoid aroma compound synthesis pathways.
Although some evidence in the literature has hinted toward a benzenoid route of synthesis in yeast (6), the low levels of intermediates and enzyme activities in this pathway in S. cerevisiae have hindered attempts to thoroughly demonstrate it. Previous studies have focused on the formation of 4-HB under aerobic conditions, searching for the aromatic ring precursor in coenzyme Q biosynthesis (21). In the current study, the production of benzenoids by H. vineae was studied under fermentation conditions to increase the formation of benzyl alcohol; this is in contrast to aerobic conditions, which stimulate the formation of aldehydes and acids. In reference to the feeding experiments, the increase in labeled 4-hydroxybenzaldehyde seen at day 12 may be explained by the observation that the Tyr pathway appears to have very slow flux to benzenoids under these conditions. We detected reduced concentrations of these compounds compared to phenylalanine-derived benzenoids, as shown in Fig. 2A, and this fact is also in agreement with previous characterizations in this yeast under fermentation conditions (20). Further studies under aerobic conditions might improve the understanding of these pathway fluxes. The blocking results with MBP inhibitor demonstrated the intracellular accumulation of the key intermediates of this pathway shown in Fig. 3. These results are in agreement with a recent report on the molecular details of the first reactions of this pathway in yeast (21), producing 4-hydroxyphenylpyruvate from Tyr. Interestingly, our results are also in partial agreement with recent molecular function predictions made through a multiomics data analysis with mass spectrometry in yeast (34). Using a metabolome containing 174 strains of S. cerevisiae, these authors observed evidence of 4-hydroxyphenylacetaldyde, 4-hydroxyphenylacetate, and 4-hydroxymandelate, among other compounds, which were predicted to be intermediates in the formation of 4-HB in yeast. However, some of the metabolites this study suggested to be involved in this pathway, such as coumarate and phenyl lactate, were not detected in our metabolic analysis of H. vineae (Fig. 3). Furthermore, transcriptomic analysis showed consistent gene expression of the eight genes that were proposed for the complete pathway presented in Fig. 4. The S. cerevisiae mutants deleted in the three key genes of the mandelate pathway proposed in this figure clearly demonstrated their involvement, although they might have other functional activities in yeast, as was shown in ARO10 (30).
Conclusions.
Taken together, these results chemically elucidate the complete pathway of benzenoid synthesis in H. vineae and molecularly validate it in S. cerevisiae. Our data demonstrate that benzenoid biosynthesis in yeasts occurs via the transamination of Phe and Tyr to their corresponding arylpyruvates, which are subsequently transformed into benzyl alcohol, and very minor concentrations of 4-hydroxybenzaldehyde, through the phenylpyruvate and mandelate pathways. The discovery of this capacity in H. vineae, along with the elucidation of the phenylpyruvate/mandelate pathway in yeast, creates new opportunities to explore the existence of this pathway in animals, as they also lack the PAL/TAL pathway. Furthermore, our results potentiate exploration of this pathway in humans, as it has recently been discovered that the human enzyme encoded by ALDH3A1 converts 4-hydroxybenzaldehyde into 4-HB in yeast cells in vivo, the benzoquinone ring of coenzyme Q10 (21). We are currently synthesizing the key enzymes of this pathway to confirm in vitro activities.
MATERIALS AND METHODS
Yeast strains and cultures.
H. vineae T02/19AF, sequenced by Giorello et al. (35), was used for the genomic, transcriptomic, and metabolic analyses, as well as for modeling the biosynthetic pathway of benzenoids. S. cerevisiae aro10Δ, S. cerevisiae dld1Δ, and S. cerevisiae dld2Δ homozygotic diploid double mutant strains from the Yeast Knockout Collection (36) were supplied by Dharmacon (Lafayette, CO). S. cerevisiae BY4743 was used as the control strain for mutant experiments. H. vineae and S. cerevisiae BY4743 strains were grown in YPD medium (2% glucose, 2% peptone, 1% yeast extract). For growth of the S. cerevisiae mutant strains, YPD medium was supplemented with 200 μg/ml gentamicin G418 (Acros-Fischer, Belgium).
Fermentation conditions.
The fermentation medium was prepared as previously described (4), with the amino acid content modified according to experimental design. All fermentations were performed with 100 mg N/liter of yeast assimilable nitrogen (YAN level, i.e., the sum of the amino acid and ammonium concentrations not including proline). The content of each amino acid, expressed in milligrams per liter, is detailed in Table 2. Experiments to quantify benzyl alcohol via Phe used 36 mg/liter isotopically marked l-phenylalanine-(phenyl-13C6) (Merck Group, Germany), while the corresponding experiments to quantify 4-hydroxybenzaldehyde used l-tyrosine-(phenyl-13C6) (Merck Group, Germany). Experiments performed on S. cerevisiae strains used media supplemented with 125 mg/liter of histidine, 500 mg/liter lysine, 150 mg/liter uracil, and 500 mg/liter leucine. Diammonium phosphate was added at a concentration of 60.30 mg/liter to equalize the YAN content in H. vineae fermentations. To maximize the production of benzenoids during S. cerevisiae fermentation, the concentration of each added amino acid was doubled instead of adding diammonium phosphate, as detailed in Table 2. The pH of each medium was adjusted with HCl to a final pH of 3.5. Equimolar concentrations of glucose and fructose were added to reach 200 g/liter, and mixed vitamins and salts were added as previously described (4). Ergosterol (10 mg/liter) was the only lipid supplement added.
TABLE 2.
Concentration of amino acids and inorganic ammonium source added to fermentation media
| Amino acid or inorganic ammonium source | Concn (mg/liter) |
|
|---|---|---|
| H. vineae | S. cerevisiae | |
| Amino acid | ||
| PRO | 60.3 | 120.6 |
| ALA | 12.1 | 24.2 |
| ARG | 90.4 | 180.8 |
| ASN | 18.1 | 36.2 |
| ASP | 42.2 | 84.4 |
| GLN | 24.1 | 48.2 |
| GLU | 60.3 | 120.6 |
| GLY | 6.0 | 12.0 |
| HIS | 18.1 | 161.2 |
| ILE | 24.1 | 48.2 |
| LEU | 36.2 | 572.4 |
| LYS | 30.1 | 560.2 |
| MET | 18.1 | 36.2 |
| PHE | 18.1 | 36.2 |
| SER | 48.2 | 96.4 |
| THR | 42.2 | 84.4 |
| TRP | 12.1 | 24.2 |
| TYR | 2.4 | 4.8 |
| VAL | 24.1 | 48.2 |
| Inorganic ammonium sourcea | ||
| DAP | 60.3 | 0 |
For S. cerevisiae, diammonium phosphate (DAP) was omitted from the medium to improve benzenoid synthesis.
Inocula were prepared in the same fermentation medium, containing 100 mg N/liter of YAN and the same specific amino acid concentrations previously detailed. Erlenmeyer flasks were incubated for 12 h in a rotary shaker at 150 rpm and 25°C. Fermentations with H. vineae were carried out in 250-ml Erlenmeyer flasks with 125 ml of medium, while fermentations with S. cerevisiae, due to their reduced production of benzenoids, were conducted in 500-ml Erlenmeyer flasks with 250 ml of medium to facilitate detection. All the fermentations were closed with cotton plugs to simulate microaerobic conditions (37). The inoculum size was 1 × 105 cells/ml in the final medium for all strains. Static batch fermentation conditions were conducted at 20°C, and each experiment was performed in triplicate. Fermentation activity was measured as CO2 weight loss and expressed in grams per 100 ml.
Samples were taken for GC-MS analyses following either 6 or 12 days of the fermentation process. The entire volume of each flask was subjected to centrifugation at 38,000 rpm for 10 min to separate the cells from the extracellular medium, after which the extracellular medium was filtered using 0.45-μm-pore membranes.
Enzymatic analysis.
Yeast extracts were obtained by collecting cells from the media of fermentations performed with H. vineae after 6 days, as described above. The fermentation medium was discarded, and the cells were resuspended in 5 ml of Tris-HCl buffer at pH 8.8 before being lysed by sonication in a Sonics Vibra-Cell (Sonics and Materials Inc., Newtown, CT). Tannat grape (V. vinifera) extract was used as a positive control; grape samples were harvested 2 weeks prior to veraison, coinciding with the highest PAL activity in red grape varieties as measured through the production of cinnamic acids (38). These extracts were used for the enzymatic analysis of PAL and TAL activity using the method described by Kalghatgi and Rao (39). Cinnamic and coumaric acids were analyzed by GC-MS following incubation of these grape and yeast extracts with the added Phe and Tyr.
Synthesis of MBP and blocking experiments.
MBP was synthesized according to the method of Brandt et al. (29). Briefly, 4.75 ml of trimethylphosphite (5 g, 40.3 mmol) was added in a dropwise fashion to a stirred volume of benzoyl chloride (5.66 g, 40.3 mmol) under nitrogen at 0°C. After the addition was completed, the reaction mixture was stirred for 30 min at 0°C and then allowed to warm to room temperature until no benzoyl chloride was observable using thin-layer chromatography. The resulting yellowish oil was purified in a chromatography column, yielding 7.1 g (82%), 1H NMR (400 MHz, CDCl3, 0.5% TMS) δ 8.24 (d, J = 7.5 Hz, 2-H), 7.53 (t, J = 7.5 Hz, 1-H), 7.494 (t, J = 7.5 Hz, 2-H), 3.910 (d, J = 10.5 Hz, 6×H). Dimethyl benzoylphosphonate (7.1 g, 33.2 mmol) was added to a solution containing 14.9 g (99.47 mmol) dry sodium iodide in 150 ml of dry acetone under nitrogen at 25°C. Solutions were stirred overnight, after which the reaction mixture was evaporated to dryness. The solid mixture was triturated several times with ethyl acetate, and the solvent was evaporated to dryness. The crude product was recrystallized using absolute ethanol to yield 5.1 g (70%), 1H NMR (500 MHz, D2O-DSS) δ 8.10 (d, J = 7.5 Hz, 2-H), 7.61 (t, J = 7.5 Hz, 1-H), 7.50 (t, J = 7.5 Hz, 2-H), 3.55 (d, J = 5.5 Hz, 3-H). The nuclear magnetic resonance (NMR) spectrum of methyl benzoylphosphonate (MBP) was confirmed after the oxidation reaction, and phosphonate signals were located in the 3.6-ppm region, shifting 0.3 ppm from starting material as published by Brandt et al. (26). For the inhibition experiments, MBP was added to the medium at the beginning of the fermentation process to achieve a final concentration of 160 μg/liter.
GC-MS analysis of intracellular and medium compounds.
The pellets resulting from centrifugation were resuspended in 5 ml of Tris-HCl buffer at pH 8.2, and cells were lysed by sonication in a Sonics Vibra-Cell (Sonics and Materials Inc., Newtown, CT). Organic compounds were extracted from the intracellular content using ethyl acetate and concentrated to dryness. Before GC-MS analysis, extracts were subjected to methylation in diethyl ether using N-nitroso-N-methylurea (NMU) in an alkaline medium. Extracellular volatile compounds were separated by liquid-liquid extraction using dichloromethane and then concentrated at 40°C under nitrogen flow. Treatment of the samples and GC-MS analysis were performed, as previously described by Martin et al. (20), using a Shimadzu-QP 2020 ULTRA mass spectrometer (Shimadzu Corp., Japan) equipped with a Stabilwax capillary column (30 m by 0.25 mm inner diameter, 0.25 μm film thickness; Restek, State College, PA).
Identification and quantification.
The analyzed aromatic compounds were identified by comparing their linear retention indices using pure standards for benzyl alcohol, 4-hydroxybenzaldehyde, mandelic acid, 4-hydroxymandelic acid, phenylacetic acid, 4-hydroxyphenylacetic acid, and phenylpyruvic acid. Mass spectrum fragmentation patterns were also compared with those stored on commercial (40–42) and our own databases. The GC-MS instrumental conditions used for quantitative purposes were as previously described (20). Isotopically labeled compounds were quantified by ion fragment comparison. Benzoic acid was used as the internal standard for intracellular samples, while 1-octanol was used for extracellular media.
Genomic and transcriptomic analyses.
The genomic sequence of H. vineae T02/19AF (35) was used to predict the genes involved in the phenylpyruvate/mandelate pathway and compared with those of S. cerevisiae available in GenBank. Transcriptomic analysis was performed using previous data obtained from H. vineae T02/19AF samples taken on days 1, 4, and 10 of fermentation (23). Briefly, total RNA was purified from three independent replicates using the RiboPure RNA purification kit for yeast (Life Technologies, Grand Island, NY, USA). The poly(A) mRNA fraction then was isolated using the Oligotex mRNA minikit (Qiagen, Hilden, Germany) and converted to indexed transcriptome sequencing (RNA-seq) libraries with the ScriptSeq v2 RNA-seq library preparation kit (Epicentre Biotechnologies, Madison, WI, USA). The nine samples were paired-end sequenced using Illumina MiSeq. Trinity was used to assemble the raw reads from transcriptomic analysis. The alignments were carried out using reciprocal BLASTx (E value cutoff of 1e−10). Transcriptomic reads were aligned against the transcriptomic reference implementing RSEM (default settings) (43).
Statistical analysis.
Analysis of variance (ANOVA) and post hoc comparisons with Tukey’s test, using a significance value of 95%, were performed on benzyl alcohol and 4-hydroxybenzaldehyde concentrations resulting from fermentation with H. vineae and S. cerevisiae strains. ANOVA comparisons were performed using STATISTICA 7.0 software (StatSoft, Tulsa, OK). Differences in mean concentrations of benzenoid compounds were evaluated using the least significant differences test.
Data availability.
Transcriptomic data are available at https://doi.org/10.6084/m9.figshare.12493556.
ACKNOWLEDGMENTS
We thank Cathy F. Clarke for her critical review of the manuscript.
We thank the following agencies for financial support: CSIC Group Project numbers 802 and 983 of UdelaR, Uruguay, Agencia Nacional de Investigación e Innovación (ANII), Hanseniaspora vineae FMV 6956 project and postdoctoral fellowship (PD_NAC_2016_1_133945), and a grant to R. Radi (“Espacio Interdisciplinario, Proyecto Centros 2015”). We also thank ANII and Lage y Cia-Lallemand Inc., Uruguay, for supporting our cooperation project on H. vineae metabolic characterization (ALI_2_2019_1_155314).
REFERENCES
- 1.Widhalm JR, Dudareva N. 2015. A familiar ring to it: biosynthesis of plant benzoic acids. Mol Plant 8:83–97. doi: 10.1016/j.molp.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Scognamiglio J, Jones L, Vitale D, Letizia CS, Api AM. 2012. Fragrance material review on benzyl alcohol. Food Chem Toxicol 50:S140–S160. doi: 10.1016/j.fct.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 3.Chen H, Fink GR. 2006. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev 20:1150–1161. doi: 10.1101/gad.1411806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrau FM, Medina K, Farina L, Boido E, Henschke PA, Dellacassa E. 2008. Production of fermentation aroma compounds by Saccharomyces cerevisiae wine yeasts: effects of yeast assimilable nitrogen on two model strains. FEMS Yeast Res 8:1196–1207. doi: 10.1111/j.1567-1364.2008.00412.x. [DOI] [PubMed] [Google Scholar]
- 5.Guillen F, Martinez AT, Martinez MJ. 1992. Substrate specificity and properties of the aryl-alcohol oxidase from the ligninolytic fungus Pleurotus eryngii. Eur J Biochem 209:603–611. doi: 10.1111/j.1432-1033.1992.tb17326.x. [DOI] [PubMed] [Google Scholar]
- 6.Clarke CF. 2000. New advances in coenzyme Q biosynthesis. Protoplasma 213:134–147. doi: 10.1007/BF01282151. [DOI] [Google Scholar]
- 7.Olson RE, Rudney H. 1983. Biosynthesis of ubiquinone. Vitam Horm 40:1–43. doi: 10.1016/s0083-6729(08)60431-8. [DOI] [PubMed] [Google Scholar]
- 8.Pierrel F, Hamelin O, Douki T, Kieffer-Jaquinod S, Mühlenhoff U, Ozeir M, Lill R, Fontecave M. 2010. Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme q biosynthesis. Chem Biol 17:449–459. doi: 10.1016/j.chembiol.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 9.Awad AM, Bradley MC, Fernández-del-Río L, Nag A, Tsui HS, Clarke CF. 2018. Coenzyme Q10 deficiencies: pathways in yeast and humans. Essays Biochem 62:361–376. doi: 10.1042/EBC20170106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsui HS, Clarke CF. 2019. Ubiquinone biosynthetic complexes in prokaryotes and eukaryotes. Cell Chem Biol 26:465–467. doi: 10.1016/j.chembiol.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández-del-Río L, Nag A, Gutiérrez Casado E, Ariza J, Awad AM, Joseph AI, Kwon O, Verdin E, de Cabo R, Schneider C, Torres JZ, Burón MI, Clarke CF, Villalba JM. 2017. Kaempferol increases levels of coenzyme Q in kidney cells and serves as a biosynthetic ring precursor. Free Radic Biol Med 110:176–187. doi: 10.1016/j.freeradbiomed.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Booth A, Masri M, Robbins D, Emerson O, Jones F, DeEds F. 1960. Urinary phenolic acid metabolities of tyrosine. J Biol Chem 235:2649–2652. [Google Scholar]
- 13.Bentley R, Ramsey VG, Springer CM, Dialameh GH, Olson RE. 1961. The origin of the benzoquinone ring of coenzyme Q9 in the rat. Biochem Biophys Res Commun 5:443–446. doi: 10.1016/0006-291X(61)90092-4. [DOI] [Google Scholar]
- 14.Hyun MW, Yun YH, Kim JY, Kim SH. 2011. Fungal and plant phenylalanine ammonia-lyase. Mycobiology 39:257–265. doi: 10.5941/MYCO.2011.39.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qualley AV, Widhalm JR, Adebesin F, Kish CM, Dudareva N. 2012. Completion of the core β-oxidative pathway of benzoic acid biosynthesis in plants. Proc Natl Acad Sci U S A 109:16383–16388. doi: 10.1073/pnas.1211001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Block A, Widhalm JR, Fatihi A, Cahoon RE, Wamboldt Y, Elowsky C, Mackenzie SA, Cahoon EB, Chapple C, Dudareva N, Basset GJ. 2014. The origin and biosynthesis of the benzenoid moiety of ubiquinone (coenzyme Q) in Arabidopsis. Plant Cell 26:1938–1948. doi: 10.1105/tpc.114.125807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soubeyrand E, Johnson TS, Latimer S, Block A, Kim J, Colquhoun TA, Butelli E, Martin C, Wilson MA, Basset GJ. 2018. The peroxidative cleavage of kaempferol contributes to the biosynthesis of the benzenoid moiety of ubiquinone in plants. Plant Cell 30:2910–2921. doi: 10.1105/tpc.18.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soubeyrand E, Kelly M, Keene SA, Bernert AC, Latimer S, Johnson TS, Elowsky C, Colquhoun TA, Block AK, Basset GJ. 2019. Arabidopsis 4-coumaroyl-CoA ligase 8 contributes to the biosynthesis of the benzenoid ring of coenzyme Q in peroxisomes. Biochem J 476:3521–3532. doi: 10.1042/BCJ20190688. [DOI] [PubMed] [Google Scholar]
- 19.Matthews RT, Yang L, Browne S, Baik M, Beal MF. 1998. Coenzyme Q10 administration increases brain mitochondrial concentrations and exerts neuroprotective effects. Proc Natl Acad Sci U S A 95:8892–8897. doi: 10.1073/pnas.95.15.8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin V, Giorello F, Fariña L, Minteguiaga M, Salzman V, Boido E, Aguilar PS, Gaggero C, Dellacassa E, Mas A, Carrau F. 2016. De novo synthesis of benzenoid compounds by the yeast Hanseniaspora vineae increases the flavor diversity of wines. J Agric Food Chem 64:4574–4583. doi: 10.1021/acs.jafc.5b05442. [DOI] [PubMed] [Google Scholar]
- 21.Payet LA, Leroux M, Willison JC, Kihara A, Pelosi L, Pierrel F. 2016. Mechanistic details of early steps in coenzyme Q biosynthesis pathway in yeast. Cell Chem Biol 23:1241–1250. doi: 10.1016/j.chembiol.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Carrau F, Gaggero C, Aguilar PS. 2015. Yeast diversity and native vigor for flavor phenotypes. Trends Biotechnol 33:148–154. doi: 10.1016/j.tibtech.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Giorello F, Valera MJ, Martin V, Parada A, Salzman V, Camesasca L, Fariña L, Boido E, Medina K, Dellacassa E, Berna L, Aguilar PS, Mas A, Gaggero C, Carrau F. 2018. Genomic and transcriptomic basis of Hanseniaspora vineae’s impact on flavor diversity and wine quality. Appl Environ Microbiol 85:e01959-18. doi: 10.1128/AEM.01959-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina K, Boido E, Fariña L, Gioia O, Gomez ME, Barquet M, Gaggero C, Dellacassa E, Carrau F. 2013. Increased flavour diversity of Chardonnay wines by spontaneous fermentation and co-fermentation with Hanseniaspora vineae. Food Chem 141:2513–2521. doi: 10.1016/j.foodchem.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 25.Martin V, Valera M, Medina K, Boido E, Carrau F. 2018. Oenological impact of the Hanseniaspora/Kloeckera yeast genus on wines—a review. Fermentation 4:76. doi: 10.3390/fermentation4030076. [DOI] [Google Scholar]
- 26.Brandt GS, Kneen MM, Chakraborty S, Baykal AT, Nemeria N, Yep A, Ruby DI, Petsko GA, Kenyon GL, McLeish MJ, Jordan F, Ringe D. 2009. Snapshot of a reaction intermediate: analysis of benzoylformate decarboxylase in complex with a benzoylphosphonate inhibitor. Biochemistry 48:3247–3257. doi: 10.1021/bi801950k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lapadatescu C, Giniès C, Le Quéré J-L, Bonnarme P. 2000. Novel scheme for biosynthesis of aryl metabolites from l-phenylalanine in the fungus Bjerkandera adusta. Appl Environ Microbiol 66:1517–1522. doi: 10.1128/aem.66.4.1517-1522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orlova I, Marshall-Colón A, Schnepp J, Wood B, Varbanova M, Fridman E, Blakeslee JJ, Peer WA, Murphy AS, Rhodes D, Pichersky E, Dudareva N. 2006. Reduction of benzenoid synthesis in petunia flowers reveals multiple pathways to benzoic acid and enhancement in auxin transport. Plant Cell 18:3458–3475. doi: 10.1105/tpc.106.046227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandt GS, Nemeria N, Chakraborty S, McLeish MJ, Yep A, Kenyon GL, Petsko GA, Jordan F, Ringe D. 2008. Probing the active center of benzaldehyde lyase with substitutions and the pseudosubstrate analogue benzoylphosphonic acid methyl ester. Biochemistry 47:7734–7743. doi: 10.1021/bi8004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kneen MM, Stan R, Yep A, Tyler RP, Saehuan C, McLeish MJ. 2011. Characterization of a thiamin diphosphate-dependent phenylpyruvate decarboxylase from Saccharomyces cerevisiae. FEBS J 278:1842–1853. doi: 10.1111/j.1742-4658.2011.08103.x. [DOI] [PubMed] [Google Scholar]
- 31.Smekal O, Yasin M, Fewson CA, Reid GA, Chapman SK. 1993. L-mandelate dehydrogenase from Rhodotorula graminis: comparisons with the L-lactate dehydrogenase (flavocytochrome b2) from Saccharomyces cerevisiae. Biochem J 290:103–107. doi: 10.1042/bj2900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinclair R, Reid G, Chapman S. 1998. Re-design of Saccharomyces cerevisiae flavocytochrome b2: introduction of L-mandelate dehydrogenase activity. Biochem J 333:117–120. doi: 10.1042/bj3330117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steenwyk JL, Opulente DA, Kominek J, Shen XX, Zhou X, Labella AL, Bradley NP, Eichman BF, Čadež N, Libkind D, De Virgilio J, Hulfachor AB, Kurtzman CP, Hittinger CT, Rokas A. 2019. Extensive loss of cell-cycle and DNA repair genes in an ancient lineage of bipolar budding yeasts. PLoS Biol 17:e3000255. doi: 10.1371/journal.pbio.3000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stefely JA, Kwiecien NW, Freiberger EC, Richards AL, Jochem A, Rush MJP, Ulbrich A, Robinson KP, Hutchins PD, Veling MT, Guo X, Kemmerer ZA, Connors KJ, Trujillo EA, Sokol J, Marx H, Westphall MS, Hebert AS, Pagliarini DJ, Coon JJ. 2016. Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat Biotechnol 34:1191–1197. doi: 10.1038/nbt.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giorello FM, Berná L, Greif G, Camesasca L, Salzman V, Medina K, Robello C, Gaggero C, Aguilar PS, Carrau F. 2014. Genome sequence of the native apiculate wine yeast Hanseniaspora vineae T02/19AF. Genome Announc 2:e00530-14. doi: 10.1128/genomeA.00530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Güldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kötter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 37.Fariña L, Medina K, Urruty M, Boido E, Dellacassa E, Carrau F. 2012. Redox effect on volatile compound formation in wine during fermentation by Saccharomyces cerevisiae. Food Chem 134:933–939. doi: 10.1016/j.foodchem.2012.02.209. [DOI] [PubMed] [Google Scholar]
- 38.Kataoka I, Kubo Y, Sugiura A, Tomana T. 1983. Changes in l-phenylalanine ammonia-lyase activity and anthocyanin synthesis during berry ripening of three grape cultivars. Engei Gakkai Zasshi 52:273–279. doi: 10.2503/jjshs.52.273. [DOI] [Google Scholar]
- 39.Kalghatgi KK, Rao P. 1975. Microbial L phenylalanine ammonia lyase. Purification, subunit structure and kinetic properties of the enzyme from Rhizoctonia solani. Biochem J 149:65–72. doi: 10.1042/bj1490065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLafferty FW. 2009. Registry of mass spectral data combined with NIST/EPA/NIH database 2008. Wiley-Blackwell, Hoboken, NJ. [Google Scholar]
- 41.Adams RP. 2007. Identification of essential oil components by gas chromatograpy/mass spectrometry, 4th ed Allured Publishing Corporation, Carol Stream, IL. [Google Scholar]
- 42.Mondello L. 2015. Mass spectra of flavors and fragrances of natural and synthetic compounds. Wiley-Blackwell, Hoboken, NJ. [Google Scholar]
- 43.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uchiyama K, Kawaguchi K, Tochikura T, Ogata K. 1969. Metabolism of aromatic amino acids in microorganisms. Part III. Metabolism of cinnamic acid in Rhodotorula. Agric Biol Chem 33:755–763. doi: 10.1080/00021369.1969.10859378. [DOI] [Google Scholar]
- 45.Zhou L, Wang J-Y, Wu J, Wang J, Poplawsky A, Lin S, Zhu B, Chang C, Zhou T, Zhang L-H, He Y-W. 2013. The diffusible factor synthase XanB2 is a bifunctional chorismatase that links the shikimate pathway to ubiquinone and xanthomonadins biosynthetic pathways. Mol Microbiol 87:80–93. doi: 10.1111/mmi.12084. [DOI] [PubMed] [Google Scholar]
- 46.Kennedy SI, Fewson CA. 1968. Enzymes of the mandelate pathway in bacterium N.C.I.B. 8250. Biochem J 107:497–506. doi: 10.1042/bj1070497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heider J, Fuchs G. 1997. Anaerobic metabolism of aromatic compounds. Eur J Biochem 243:577–596. doi: 10.1111/j.1432-1033.1997.00577.x. [DOI] [PubMed] [Google Scholar]
- 48.Farhi M, Dudareva N, Masci T, Weiss D, Vainstein A, Abeliovich H. 2006. Synthesis of the food flavoring methyl benzoate by genetically engineered Saccharomyces cerevisiae. J Biotechnol 122:307–315. doi: 10.1016/j.jbiotec.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Long A, Ward OP. 1989. Biotransformation of benzaldehyde by Saccharomyces cerevisiae: characterization of the fermentation and toxicity effects of substrates and products. Biotechnol Bioeng 34:933–941. doi: 10.1002/bit.260340708. [DOI] [PubMed] [Google Scholar]
- 50.Long MC, Nagegowda DA, Kaminaga Y, Ho KK, Kish CM, Schnepp J, Sherman D, Weiner H, Rhodes D, Dudareva N. 2009. Involvement of snapdragon benzaldehyde dehydrogenase in benzoic acid biosynthesis. Plant J 59:256–265. doi: 10.1111/j.1365-313X.2009.03864.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Transcriptomic data are available at https://doi.org/10.6084/m9.figshare.12493556.