Sanchez-Huertas et al. demonstrate that the +TIP Navigator-1 (NAV1) couples F-actin and microtubules in the growth cone of cortical axons. This property enables NAV1 to promote microtubule persistence in the growth cone periphery and controls growth cone dynamics and steering.
Abstract
Microtubule (MT) plus-end tracking proteins (+TIPs) are central players in the coordination between the MT and actin cytoskeletons in growth cones (GCs) during axon guidance. The +TIP Navigator-1 (NAV1) is expressed in the developing nervous system, yet its neuronal functions remain poorly elucidated. Here, we report that NAV1 controls the dynamics and motility of the axonal GCs of cortical neurons in an EB1-dependent manner and is required for axon turning toward a gradient of netrin-1. NAV1 accumulates in F-actin–rich domains of GCs and binds actin filaments in vitro. NAV1 can also bind MTs independently of EB1 in vitro and crosslinks nonpolymerizing MT plus ends to actin filaments in axonal GCs, preventing MT depolymerization in F-actin–rich areas. Together, our findings pinpoint NAV1 as a key player in the actin–MT crosstalk that promotes MT persistence at the GC periphery and regulates GC steering. Additionally, we present data assigning to NAV1 an important role in the radial migration of cortical projection neurons in vivo.
Introduction
During development of the nervous system, neurons project their axons over long distances to innervate specific targets. Axon guidance is driven by the growth cone (GC), which senses and integrates extrinsic cues at the tip of the axon and translates them into cytoskeletal changes that produce traction and steering (Dent et al., 2011; Kolodkin and Tessier-Lavigne, 2011). The peripheral domain (P-domain) of the GC concentrates the actin network which is organized as a branched meshwork or dense bundles in the lamellipodia and filopodia, respectively. Microtubules (MTs) concentrate into the central domain, where they are constrained by actomyosin contractile structures sitting at the transition zone (Dent et al., 2011; Geraldo and Gordon-Weeks, 2009). Only a few dynamic MTs, referred to as “pioneer” MTs, cross the transition zone and invade the actin-rich periphery of the GC. This process has been shown to be determinant for GC turning and outgrowth in response to external cues (Liu and Dwyer, 2014; Schaefer et al., 2008). Numerous studies have established that the crosstalk between MTs and F-actin in the GC is crucial for axon guidance, and members of the MT plus-end tracking protein (+TIP) family have pivotal roles in mediating this crosstalk. For instance, the spectraplakin ACF7/MACF1, the adenomatous polyposis coli (APC) protein, cytoplasmic linker protein (CLIP)-associating proteins (CLASPs), and XMAP215/Colonic and hepatic tumor overexpressed gene (Ch-TOG) are actin-binding +TIPs that can crosslink MTs and actin fibers in the GC (Coles and Bradke, 2015; Cammarata et al., 2016). This coupling has been proposed to guide the growth of MTs along preexisting F-actin bundles and control their dynamics (Hur et al., 2011; Koester et al., 2007; Purro et al., 2008; Slater et al., 2019). Besides, mounting evidence in nonneuronal cells also suggests that actin remodeling can be influenced by MT plus ends through APC or CLIP170 via their interaction with the formin mDia1 (Henty-Ridilla et al., 2016; Okada et al., 2010).
Neuron Navigator (NAV) proteins are a family of large multidomain +TIPs that share several conserved protein domains including a calponin-homology (CH) domain in the N-terminus, several coiled-coil regions, and an AAA+ (ATPases associated with various cellular activities) domain in the C-terminus (Maes et al., 2002; Stringham et al., 2002). Tandem CH domains have been shown to confer both actin and MT binding to a variety of proteins, and domains of the AAA+ family are found in a large number of proteins involved in protein degradation, DNA replication, MT motor movement, and MT severing (Stradal et al., 1998; White and Lauring, 2007). The evolutionary conserved NAV family comprises Caenorhabditis elegans UNC-53, Drosophila melanogaster Sickie, and three vertebrate proteins named NAV1, NAV2, and NAV3. UNC-53 has been shown to control cell migration and axon outgrowth and guidance in the worm, and Drosophila Sickie has been implicated in the axonal outgrowth of mushroom body neurons (Abe et al., 2014; Hekimi and Kershaw, 1993; Stringham et al., 2002; Stringham and Schmidt, 2009). In these invertebrates, NAVs have been suggested to participate in signaling pathways controlling actin cytoskeleton remodeling (Abe et al., 2014; Marcus-Gueret et al., 2012; Pandey et al., 2018; Schmidt et al., 2009).
The three vertebrate NAV proteins are also expressed in the nervous system (Stringham and Schmidt, 2009), with NAV2 being the closest orthologue of UNC-53 and Sickie. NAV2 is able to rescue the axonal elongation defects of C. elegans unc-53 mutants, participates in neurite outgrowth, and plays a prominent role in brain development (McNeill et al., 2011, 2010; Merrill et al., 2002; Muley et al., 2008; Peeters et al., 2004). NAV3 has been proposed to be involved in neuron growth and regeneration (Coy et al., 2002). In rodents, the expression of the NAV1 gene is largely restricted to the developing nervous system, and the NAV1 protein appears to be specifically enriched at neuritic tips and axonal GCs (Martínez-López et al., 2005; van Haren et al., 2014). In contrast to NAV2 and NAV3, NAV1 does not possess a CH domain, which could account for distinct cellular activities (Maes et al., 2002). NAV1 was reported to interact with the RhoGEF Trio at MT plus ends, enhancing Trio-induced Rac1 activation and neurite outgrowth (van Haren et al., 2014). NAV1 has also been proposed to be required for hindbrain neuron migration toward a netrin-1 gradient ex vivo (Martínez-López et al., 2005). Nevertheless, the functions of NAV1 and the molecular mechanisms whereby it operates in neurons remain poorly elucidated.
Here, we show that NAV1 is an end-binding protein (EB)-dependent +TIP that stabilizes nonpolymerizing MT plus ends in F-actin–rich domains and crosslinks them to actin filaments in the GC periphery of cortical neurons, stabilizing MTs in F-actin–rich domains. We provide evidence that this mechanism is determinant for GC dynamics and steering. We also uncover NAV1 as a downstream effector of netrin-1–induced chemoattraction in the GC and a regulator of neuronal radial migration during corticogenesis.
Results
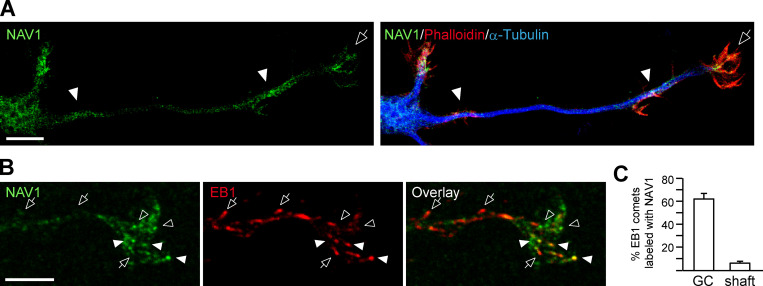
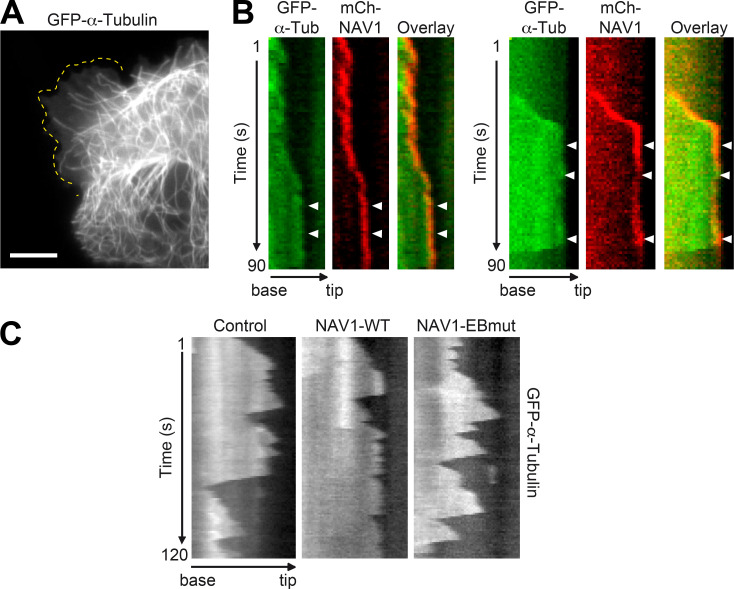
NAV1 controls GC dynamics and axon bending
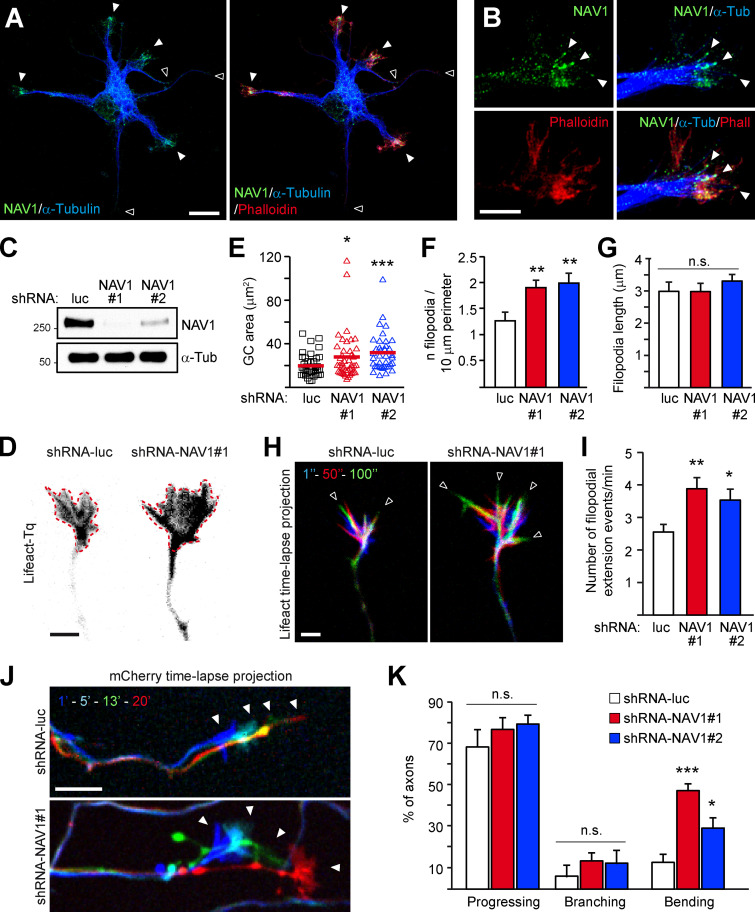
We and others have shown that NAV1 is enriched at neurite tips and GCs of hippocampal neurons in culture (Martinez-Lopez et al., 2005; van Haren et al., 2014). To characterize in detail the localization of NAV1 in GCs, we labeled endogenous NAV1, actin fibers, and MTs in cultured cortical neurons. In unpolarized neurons, we found that NAV1 is enriched specifically in enlarged neurite tips that display a high F-actin content (Fig. 1 A). In polarized neurons, NAV1 is enriched in F-actin–rich areas inside axonal or dendritic GCs and even along the axon shaft (Fig. S1 A). High-resolution microscopy images of axonal GCs revealed the presence of endogenous NAV1 at the tip of MTs that are embedded within the peripheral F-actin–rich subdomain (Fig. 1 B). Consistent with this observation, ∼62% of the EB1-positive MT plus ends within the P-domain of GCs were colabeled with NAV1 compared with only ∼6% along the actin-free axon shaft (Fig. S1, B and C). This analysis also revealed an EB1-negative granular pattern of NAV1 in the GCs (Fig. S1 B), which we previously observed in GCs of hippocampal neurons (van Haren et al., 2014). Given the prominent localization of NAV1 in GCs, we investigated the effect of the depletion of NAV1 on GC morphology and dynamics. To this end, we cotransfected cultured cortical neurons with NAV1-targeting shRNAs (Fig. 1 C) and the F-actin reporter Lifeact-Tq (Riedl et al., 2008) and we monitored the dynamics of their axonal GCs. Interestingly, we measured larger GCs displaying a higher density of filopodia in NAV1-depleted neurons (Fig. 1, D–F), with no change in the average filopodia length (Fig. 1 G). Time-lapse analysis revealed that NAV1 depletion boosted GC dynamics, measured as an increase in the frequency of filopodial extension events (Fig. 1, H and I; and Video 1). Conversely, the overexpression of GFP-NAV1 in cortical neurons led to smaller and less dynamic axonal GCs (Fig. S2). Next, we investigated whether NAV1 depletion had further consequences on the motility of axonal GCs. We observed that the depletion of NAV1 did not affect their protrusive motility (progress; Fig. 1, J and K). Among the GCs that progressed, we did not observe differences in the frequency of GC branching either, but we found that NAV1 depletion increased the frequency of spontaneous axon bending (Fig. 1, J and K; and Video 2). Together, these data show that NAV1 attenuates GC spreading and influences axon steering.
Figure 1.
NAV1 controls GC dynamics and axon bending. (A and B) Immunostaining of NAV1, MTs (α-tubulin), and F-actin (phalloidin) in E14.5 cultured cortical neurons (2 DIV). (A) Closed arrowheads mark neurite tips enriched with F-actin and NAV1. Open arrowheads mark neurite tips devoid of F-actin and NAV1. Scale bar, 10 µm. See also Fig. S1 A. (B) High-resolution images of an axonal GC. Arrowheads point to MT plus ends embedded in F-actin and decorated with NAV1. Scale bar, 5 µm. See also Fig. S1 B. Results were replicated in 30 neurons and 12 GCs from two independent cultures. (C) Immunoblots of lysates of cultured cortical neurons (3 DIV) transduced with the indicated shRNAs by lentiviral infection at 0 DIV. α-tubulin was used as a loading control. (D–I) Axonal GCs of cortical neurons (3 DIV) transfected with Lifeact-Tq and the indicated shRNAs and imaged by time-lapse microscopy during 2 min. (D) Lifeact-Tq images of GCs of control and NAV1-depleted neurons. Dashed red lines outline the GC areas. Scale bar, 2 µm. See also Video 1. (E–G) Quantifications of GC area (E), density of filopodia (F), and average filopodia length (G). See also Fig. S2. (H) Color-coded time-lapse projections of the Lifeact-Tq signal. The indicated time points were used. Arrowheads mark filopodia. Scale bar, 2 µm; time, seconds. See also Video 1. (I) GC dynamics of control and NAV1-depleted neurons. n = 39 (shRNA-luc), 43 (shRNA-NAV1#1), and 38 (shRNA-NAV1#2) GCs from four independent experiments. (J and K) Axonal GCs of cortical neurons (3 DIV) transfected with the indicated shRNAs and imaged by time-lapse microscopy for 20 min. (J) Color-coded time-lapse projections of the mCherry reporter in GCs. The indicated time points were used. Arrowheads mark positions of the GCs. Scale bar, 10 µm; time, minutes. See also Video 2. (K) Proportion of axons showing net progression, branching, or bending. Only the axons showing net progression were scored as branching or bending. n = 61 (shRNA-luc), 72 (shRNA-NAV1#1) and 61 (shRNA-NAV1#2) axons from four independent experiments. All bar histograms show means ± SEM. Dot plot shows means. Analyzed by two-tailed Student's t test. n.s., nonsignificant.
Figure S1.
NAV1 accumulates in F-actin–rich areas in cortical neurons and colocalizes with EB1 at MT plus ends in the GC. Related to Fig. 1. (A and B) Confocal microscopy images of GCs of cortical neurons (2 DIV) stained with the indicated antibodies and phalloidin-TRITC. Results were replicated in multiple neuronsfrom two independent cultures. (A) The arrow points to an axonal GC enriched with F-actin and NAV1. Arrowheads mark areas along the axon shaft enriched with F-actin and NAV1. Scale bar, 10 µm. (B) Arrows mark EB3-postive/NAV1-negative MT plus ends. Closed arrowheads mark EB3/NAV1-positive MT plus ends. Open arrowheads mark EB3-negative NAV1 granules. Scale bar, 5 µm. (C) Percentage of EB1 comets colabeled with NAV1 in GCs and 10 µm upstream in their axon shafts from B. n = 214 EB1 comets in 17 GCs from two independent experiments. Histogram shows means ± SEM.
Video 1.
Axonal GCs of NAV1-depleted neurons are larger and more dynamic than those of control neurons. Related to Fig. 1, D–I. Time-lapse microscopy of neurons transfected with Lifeact-Tq and the indicated shRNAs. Time interval: 1 second.
Figure S2.
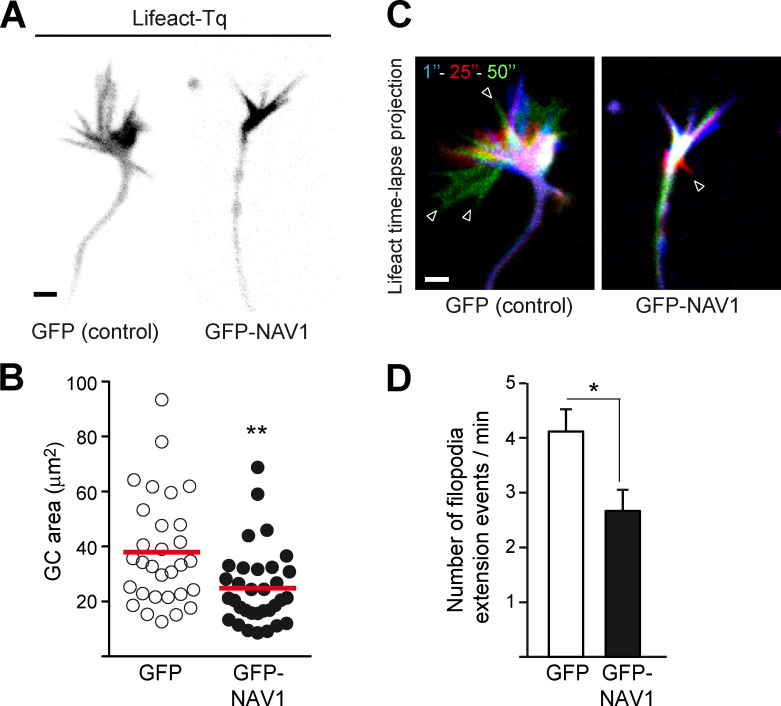
Overexpression of NAV1 compacts the GC and inhibits its dynamics. Related to Fig. 1. Axonal GCs of cortical neurons (2 DIV) transfected with Lifeact-Tq and GFP or GFP-NAV1, and imaged by time-lapse microscopy during 2 min. (A) Lifeact-Tq images of GCs of control and NAV1-overexpressing neurons. Scale bar, 2 µm. (B) Quantification of GC area. (C) Color-coded time-lapse projections of the Lifeact-Tq signal. The indicated time points were used. Arrowheads mark filopodia. Scale bar, 2 µm; time, seconds. (D) GC dynamics in control and NAV1-overexpressing neurons. n = 30 (GFP) and 33 (GFP-NAV1) GCs from three independent experiments. Dot plot shows means. Histogram shows means ± SEM. Analyzed by two-tailed Student's t test.
Video 2.
Axons of NAV1-depleted neurons show more frequent bending and changes of direction than those of control neurons. Related to Fig. 1, J and K. Time-lapse microscopy of neurons transfected with the indicated shRNAs with a mCherry reporter. Time interval: 1 minute.
NAV1 mediates netrin-1–evoked GC turning
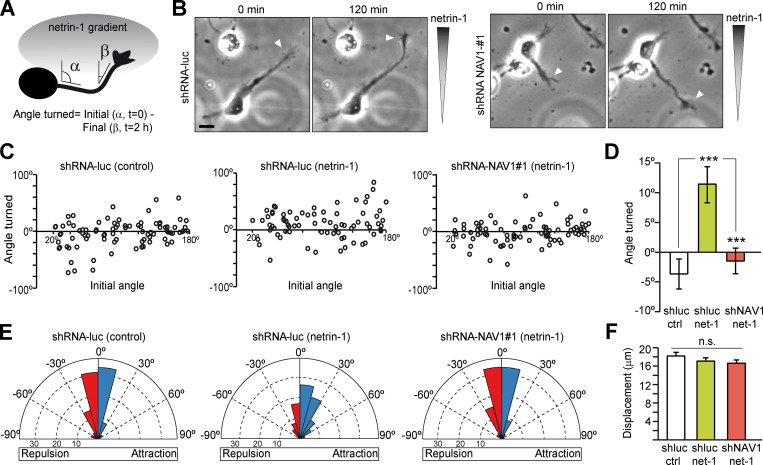
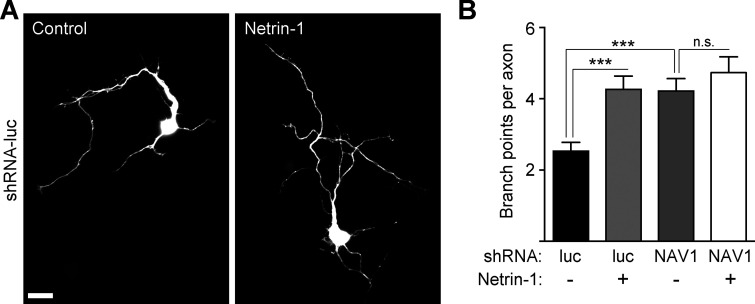
Since NAV1 depletion increased the frequency of spontaneous steering of GCs, we sought to investigate whether NAV1 participates to axon guidance signaling. Netrin-1 acts as a chemoattractant in early stage cortical neuron cultures, and NAV1 is required for hindbrain neuron migration toward a netrin-1 gradient ex vivo (DeGeer et al., 2015; Martínez-López et al., 2005). To assess NAV1 participation in netrin-1–dependent axon turning, we analyzed the effect of NAV1 depletion on neurons exposed to a gradient of netrin-1 in a Dunn chamber (Yam et al., 2009). Cortical neuron cultures were infected with lentiviruses carrying control or NAV1-targeting shRNAs and exposed to netrin-1, and their axons were monitored by time-lapse imaging for 2 h. The angle turned by each axon was defined by the initial (α) and final (β) angles of the distal segment of the axon respective to a line parallel to the gradient, in a way that positive angles mean axon turning toward the gradient (Fig. 2 A). Regardless of their initial angle, the axons of control neurons exposed to netrin-1 displayed more frequent positive turning angles compared with those of control neurons exposed to BSA. In the rose diagrams, this was reflected by a biased distribution of turned angles toward positive values. In contrast, the distribution of turned angles in NAV1-depleted neurons exposed to netrin-1 appeared to be randomized (Fig. 2, B–E; and Video 3). Besides, we found no differences in axon extension among all conditions during the imaging time (Fig. 2 F), indicating that the disruption of netrin-1 attractiveness found in NAV1-deficient axons was not due to reduced outgrowth. Since netrin-1 also stimulates axon branching in cortical neurons (Dent et al., 2004), we assessed whether NAV1 mediates this effect. Interestingly, NAV1-depleted cortical neurons per se displayed increased axon branching to the same extend as control neurons exposed to netrin-1 for 24 h, indicating that NAV1 participates in the control of this process (Fig. S3). However, NAV1-depleted neurons did not show any further increase in axon branching upon netrin-1 treatment (Fig. S3). From these data, we conclude that NAV1 is necessary for netrin-1–dependent GC turning and axon branching.
Figure 2.
NAV1 mediates netrin-1–evoked GC turning. Cortical neurons were transduced with the indicated shRNAs by lentiviral infection at 0 DIV and imaged at 2 DIV by phase-contrast microscopy for 2 h while being exposed to a gradient of 200 ng/ml netrin-1 in a Dunn chamber. (A) The angle turned by each axon was defined by the initial (α) and final (β) angles of the distal segment of the axon respective to a line parallel to the gradient. Angle turned >0° indicates attraction, and <0° indicates repulsion. (B) Control and NAV1-depleted neurons in the Dunn chamber imaged at the beginning (0 min) and end (120 min) of the assay. Arrowheads point to the axon tips. Netrin-1 concentration increases along the vertical axis of images. Scale bar, 20 µm. See also Video 3. (C) Scatter plot of the angle turned compared with the initial angle of axons for initial angles >20°. (D) Mean angle turned by axons analyzed in C. (E) Rose histograms depicting the distribution of angles turned by axons. Angles were clustered in bins of 15°, and the proportion of axons per bin is represented by the radius of each segment. Blue bins indicate attraction, and red bins indicate repulsion. (F) Axon length of neurons from C. n = 87 (shRNA-luc/BSA), 76 (shRNA-luc/netrin-1), and 80 (shRNA-NAV1#1/netrin-1) axons from three independent experiments. Bar histograms show means ± SEM. Analyzed by two-tailed Student's t test.
Video 3.
NAV1 depletion impairs axon turning toward a netrin-1 gradient. Related to Fig. 2. Phase-contrast time-lapse microscopy of control (shRNA-luc) and NAV1-depleted (shRNA-NAV1#1) neurons exposed to netrin-1 in a Dunn chamber. The higher concentration of netrin-1 is at the top of the images. Time interval: 4 minutes.
Figure S3.
NAV1 depletion impairs netrin-1–evoked axon branching. Related to Fig. 2. Cortical neurons transfected with the indicated shRNAs at 1 DIV, treated with netrin-1 (500 ng/ml) for 24 h, and fixed at 3 DIV. (A) Anti-mCherry immunostaining of neurons transfected with shRNA-luc and either left untreated or treated with netrin-1. Scale bar, 20 µm. (B) Quantitative analysis of branch points per axon. n = 60 (shRNA-luc), 59 (shRNA-luc + netrin-1), 55 (shRNA-NAV1), and 49 (shRNA-NAV1 + netrin-1) neurons from two independent experiments. Histogram shows means ± SEM. Analyzed by two-tailed Student's t test.
NAV1 is an actin-binding +TIP that requires MT plus-end tracking to control GC dynamics
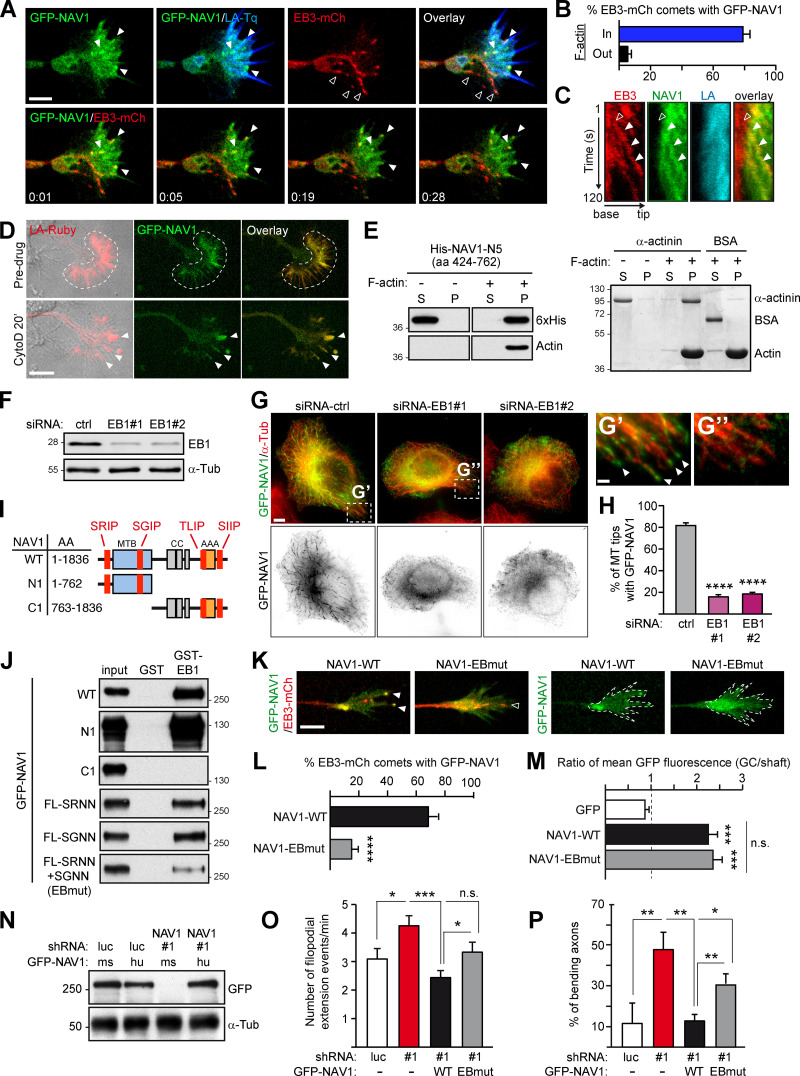
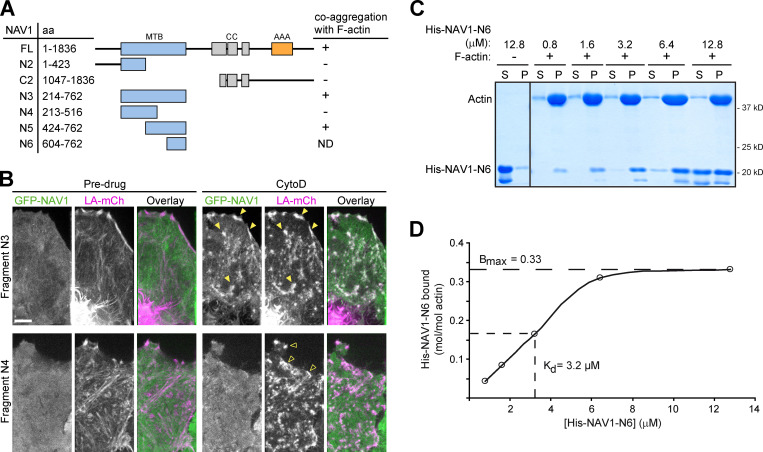
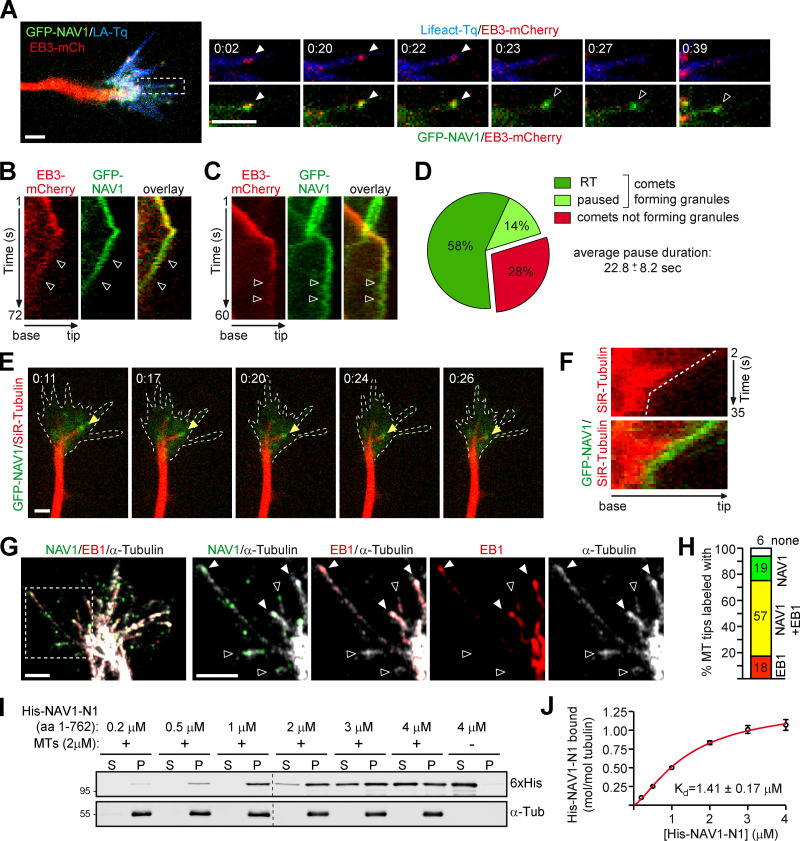
To gain insight into the mechanisms by which NAV1 regulates GC dynamics, we investigated the MT plus-end tracking dynamics of NAV1. First, we performed live-imaging microscopy on axonal GCs of cortical neurons coexpressing GFP-NAV1, Lifeact-Tq, and EB3-mCherry, which accumulates at the plus end of polymerizing MTs (Stepanova et al., 2003). Similarly to endogenous NAV1, GFP-NAV1 decorated ∼80% of the EB3-mCh–positive MT plus ends within the F-actin–rich filopodia and lamellipodia of the peripheral domain and in the transition zone of GCs, compared with only ∼5% of EB3-mCherry comets outside these areas (Fig. 3, A and B; and Video 4). Kymographs obtained from the transition zone of GCs revealed that NAV1 labels MT plus ends once they invade the F-actin–rich domains (Fig. 3 C). To assess whether NAV1 physically associates with the actin network, we examined its localization inside GCs upon disruption of the actin network organization induced by cytochalasin D (Mortensen and Larsson, 2003). We found that GFP-NAV1 relocalized together with Lifeact-positive aggregates upon cytochalasin D treatment of neurons (Fig. 3 D), suggesting that it associates directly or indirectly with the actin network. To delineate actin-binding regions within NAV1, we expressed truncated forms of GFP-NAV1 in HeLa cells and took advantage of the coaggregation feature shown by NAV1 and F-actin upon treatment with cytochalasin D. By using this assay, we defined a 339-aa NAV1 fragment (aa 424–762; NAV1-N5), which retained coaggregation with F-actin (Fig. S4, A and B). We then tested the actin-binding properties of this fragment in an in vitro F-actin cosedimentation assay. When incubated with preassembled skeletal muscle actin filaments, this fragment cosedimented with F-actin to the pellet fraction, whereas it remained in the soluble fraction in the absence of F-actin, similarly to the positive control α-actinin (Fig. 3 E). A smaller NAV1 fragment (aa 604–762; NAV1-N6) was then used to calculate its affinity (equilibrium dissociation constant, or Kd) for F-actin (Fig. S4, C and D). This revealed a low affinity (Kd ∼3.2 µM), similar to that reported before for tandem CH domains (Gimona et al., 2002). Based on our findings, we conclude that NAV1 is an F-actin-binding +TIP.
Figure 3.
NAV1 is an actin-binding +TIP that requires MT plus-end tracking to control GC dynamics. (A) Top: Axonal GC of a cortical neuron (2 DIV) cotransfected with GFP-NAV1, EB3-mCherry, and Lifeact-Tq. Bottom: Time-lapse composition of the GFP-NAV1/EB3-mCherry overlay. Closed arrowheads point to EB3/NAV1-positive MT plus ends (comets) inside F-actin. Open arrowheads mark EB3-positive/NAV1-negative MT plus ends outside F-actin. Scale bar, 5 µm; time, minutes:seconds. See also Video 4. (B) Percentage of EB3-mCherry comets decorated by GFP-NAV1 inside and outside F-actin in GCs as in A. n = 159 total EB3-mCherry comets analyzed in 28 GCs from five independent experiments. (C) Kymographs obtained from a GC from A. NAV1 (green) tip-tracks MTs (red) only inside F-actin (blue). Arrowheads mark an EB3-mCherry comet outside (open) and inside (closed) F-actin. (D) Time-lapse composition of the axonal GC of a cortical neuron (2 DIV) cotransfected with GFP-NAV1 and Lifeact-Ruby, before and after exposure to cytochalasin D (CytoD) for 20 min. Dashed lines delineate the F-actin-rich P-domain. Arrowheads point to the aggregates of F-actin and NAV1 protein after cytochalasin D treatment. Scale bar, 10 µm. Averaged Pearson´s coefficient = 0.48 ± 0.24 (before cytochalasin D); 0.66 ± 0.16 (after cytochalasin D); n = 11 GCs from two independent experiments. (E) Immunoblots of the soluble (S) and pellet (P) fractions of an in vitro F-actin cosedimentation assay with purified His-NAV1-N5 fragment (left). BSA and α-actinin were used as a negative and positive control and visualized by Coomassie staining (right). Results were replicated three times. See also Fig. S4. (F–H) Localization of GFP-NAV1 in EB1-depleted HeLa cells. (F) Immunoblots of lysates of HeLa cells transfected with the indicated siRNAs. α-Tubulin was used as a loading control. (G–G′′) HeLa cells cotransfected with the indicated siRNAs and GFP-NAV1 and stained with an anti-α-tubulin antibody. Arrowheads point to MT tips decorated with GFP-NAV1. Scale bars, 5 and 1 µm. (H) Percentage of MT tips decorated with GFP-NAV1 in cells from G. n = 246 (siRNA-ctrl), 350 (siRNA-EB1#1), and 326 (siRNA-EB1#2) MT tips from 26 (siRNA-ctrl), 20 (siRNA-EB1#1), and 20 (siRNA-EB1#2) cells from two independent experiments. (I) GFP-NAV1 constructs used in J. AAA+, predicted ATPase domain; CC, coiled coil; MTB, predicted MT-binding domain. SRIP, SGIP, TLIP, and SIIP are SxIP motifs. (J) Anti-GFP immunoblots of GST-EB1 affinity pull-down assays with the indicated truncated or mutated forms of GFP-NAV1 expressed in HEK293T cells. A GST column was used as a control. FL-SRNN+SGNN is NAV1-EBmut. Results were replicated three times. (K) Time-lapse microscopy of axonal GCs of cortical neurons (2 DIV) transfected with EB3-mCherry, Lifeact-Tq, and the indicated forms of GFP-NAV1. Dashed lines outline Lifeact-Tq regions (not depicted). Arrowheads point to EB3/NAV1-positive (closed) and EB3-positive/NAV1-negative (open) MT plus ends. Scale bar, 5 µm. See also Video 5. (L) Percentage of EB3-mCherry comets decorated with GFP-NAV1 in GCs from K. n = 69 (WT) and 82 (EBmut) EB3-mCherry comets from 31 (WT) and 35 (EBmut) GCs from three independent experiments. (M) Ratio of mean GFP fluorescence intensity between the Lifeact-delineated area and the axon shaft of GCs from K. n = 32 (GFP), 34 (WT), 36 (EBmut) GCs from three independent experiments. (N–P) Rescue experiments of NAV1-depletion phenotypes shown in Fig. 1, I and K. (N) Immunoblots of lysates of HEK293T cells cotransfected with the indicated shRNAs and GFP-NAV1 constructs. α-Tubulin was used as a loading control. Human GFP-NAV1 expression is resistant to shRNA-NAV1#1. ms, mouse; hu, human. (O) Dynamics of axonal GCs as in Fig. 1 I. n = 29 (shRNA-luc), 35 (shRNA-NAV1#1), 32 (WT rescue), and 32 (EBmut rescue) GCs from three independent experiments. (P) Percentage of axons bending as in Fig. 1 K. n = 45 (shRNA-luc), 57 (shRNA-NAV1#1), 46 (WT rescue), and 42 (EBmut rescue) axons from three independent experiments. Histograms show means ± SEM. Analyzed by two-tailed Student's t test.
Video 4.
GFP-NAV1 tracks polymerizing MT plus ends within the F-actin network in GCs. Related to Fig. 3, A and B. Time-lapse microscopy of the axonal GC of a neuron transfected with GFP-NAV1, EB3-mCherry, and Lifeact-Tq. MT plus ends polymerizing outside the F-actin network are not labeled with GFP-NAV1. Time interval: 1 second.
Figure S4.
NAV1 binds F-actin. Related to Fig. 3. (A) Schematic representation of the full-length and truncated forms of GFP-NAV1 constructs used in B. AAA+, predicted ATPase domain; CC, coiled coil; MTB, predicted microtubule-binding domain. (B) Fluorescence microscopy images of HeLa cells transfected with Lifeact-mCherry and the indicated GFP-NAV1 constructs and either left untreated or treated with cytochalasin D for 1 min. In control cells, GFP-NAV1-N3 and N4 appear soluble in the cytosol. GFP-NAV1-N3 relocalizes to Lifeact-positive aggregates of F-actin upon cytochalasin D treatment (closed arrowheads), whereas GFP-NAV1-N4 remains soluble (open arrowheads). Scale bar, 5 µm. (C) Coomassie-stained gel of the soluble (S) and pellet (P) fractions of an in vitro F-actin cosedimentation assay with purified His-tagged NAV1-N6 fragment (aa 604–762). (D) Saturation binding curve for His-NAV1-N6 and equilibrium dissociation constant (Kd) calculated as described in Materials and methods.
NAV1 is a +TIP that binds EB1 in vitro (van Haren et al., 2009, 2014). Therefore, we assessed the importance of EB1 in the localization of NAV1 at MT plus ends by cotransfecting EB1-targeting siRNAs and GFP-NAV1 in HeLa cells (Fig. 3, F and G). We found that the MT plus-end localization of NAV1 was reduced by ∼80% in EB1-depleted cells (Fig. 3, G and H), indicating that the MT plus-end tracking ability of NAV1 mostly relies on its interaction with EB1. Most EB-binding +TIPs interact with EB proteins either via a CAP-Gly domain or via Ser-x-Ile-Pro (SxIP) sequence motifs (Akhmanova and Steinmetz, 2015), and we pinpointed four SxIP motifs in the sequence of NAV1 (Fig. 3 I). To dissect the interaction between NAV1 and EB1, we tested truncated and SxIP-mutated forms of GFP-NAV1 in a pull-down assay with a GST-EB1 affinity column (Fig. 3, I and J). Both full-length NAV1 and its N-terminal half (NAV1-N1) containing two SxIP motifs interacted with EB1. Individual mutation of these motifs only had a minor effect on the interaction of full-length NAV1 with EB1, whereas the combined mutation of both SxIP motifs (SRNN+SGNN, named NAV1-EBmut) strongly impaired this interaction (Fig. 3 J). From these results, we conclude that the two SxIP motifs located in the N-terminus of NAV1 constitute the major EB1-binding sites. To assess the MT tip tracking properties of NAV1-EBmut, we coexpressed GFP-NAV1 constructs with EB3-mCherry and Lifeact-Tq in cultured neurons, and performed time-lapse microscopy analysis of axonal GCs. As expected, NAV1-EBmut decorated ∼80% less EB3-positive comets than NAV1-WT in the P-domain of GCs (Fig. 3, K and L; and Video 5). Interestingly, quantification of the mean GFP fluorescence intensity ratio between the actin-rich regions of GCs and their axonal shafts showed no difference between NAV1-WT– and NAV1-EBmut–expressing neurons, indicating that the MT plus-end tracking property of NAV1 is dispensable for its localization to the F-actin network (Fig. 3, K and M). Combined, these data suggest that NAV1 binds to F-actin, which causes its accumulation in F-actin–rich domains of GCs and thereby its preferential recruitment to MT plus ends via EB1 in these domains.
Video 5.
Point mutation of both SxIP motifs mediating NAV1 binding to EB1 impairs its MT plus-end tracking ability in F-actin–rich regions of GCs. Related to Fig. 3 K. Time-lapse microscopy of neurons expressing EB3-mCherry, Lifeact-Tq, and either GFP-NAV1-WT (top) or GFP-NAV1-EBmut (bottom). Time interval: 1 second.
We then assessed the ability of the EB1-binding–deficient mutant of NAV1 to rescue the phenotypes observed in the GCs of NAV1-depleted neurons. To do so, we coexpressed NAV1-targeting shRNAs with shRNA-resistant forms of human NAV1 in cortical neurons (Fig. 3 N) and monitored their axonal GCs as in Fig. 1, I and K. The increased GC dynamics measured in NAV1-depleted neurons was fully rescued by expression of GFP-NAV1-WT, but not as efficiently by expression of GFP-NAV1-EBmut (Fig. 3 O). Similarly, coexpression of GFP-NAV1-EBmut did not rescue the increased frequency of axon bending as efficiently as GFP-NAV1-WT (Fig. 3 P). These results demonstrate that NAV1 requires MT plus-end tracking ability to control GC dynamics and steering.
NAV1 promotes the persistence of MTs in the P-domain of GCs by preventing their depolymerization
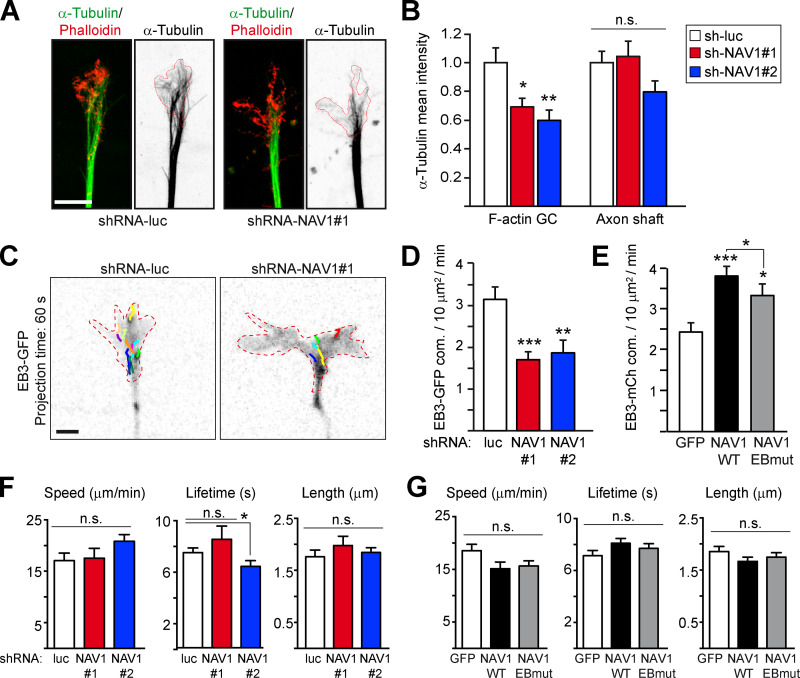
GC protrusion and steering are influenced by MT dynamics in the P-domain (Kahn and Baas, 2016), so we examined the organization of the MT cytoskeleton in the axonal GCs of NAV1-depleted neurons. Quantitative analysis of the MT content of GCs revealed that the depletion of NAV1 led to an ∼40% reduction in the density of MTs in the phalloidin-positive areas of the GC periphery, but not in the proximal axon shaft (Fig. 4, A and B). Consistent with this observation, NAV1 depletion reduced the density of EB3-GFP–labeled MT plus ends polymerizing in the F-actin–rich areas of axonal GCs (Fig. 4, C and D). Conversely, overexpression of GFP-NAV1-WT in neurons increased the density of EB3-mCherry–labeled MT tips polymerizing inside the F-actin–rich areas of their GCs. Overexpression of GFP-NAV1-EBmut also increased the density of EB3 comets, but significantly less than WT NAV1 (Fig. 4 E). These findings suggested that NAV1 may regulate MT plus-end dynamics inside F-actin–rich domains. To address this hypothesis, we measured the polymerization parameters of the EB3-GFP-labeled MT plus ends in the axonal GCs of either NAV1-depleted or NAV1-overexpressing neurons. Depletion of NAV1 did not significantly interfere with MT plus-end growth speed, lifetime, or length in the F-actin–rich GC periphery (Fig. 4 F). Likewise, the growth parameters of MTs measured in GCs of neurons overexpressing GFP-NAV1 remained unchanged (Fig. 4 G). To quantify MT dynamics in more detail, we overexpressed mCherry-NAV1 constructs in LLCPK-1α cells stably expressing GFP-α-tubulin (Rusan et al., 2001) and performed total internal reflection fluorescence (TIRF) time-lapse microscopy in the vicinity of the basal actin cortex (Fig. S5 A). This confirmed that mCh-NAV1-WT, but not mCh-NAV1-EBmut, tip-tracks dynamic MT plus ends in LLCPK-1α cells (Video 6) and also revealed that mCherry-NAV1 persists at the MT plus ends that occasionally pause underneath the plasma membrane (Fig. S5 B). Kymograph analysis of MTs exploring actin-rich lamellipodia revealed that their growth parameters were similar in control and NAV1-expressing cells (Table 1), corroborating our data obtained in cultured neurons. By contrast, the mean duration of MT depolymerization events was shorter in cells expressing NAV1-WT, and the MTs spent more time pausing (Table 1 and Fig. S5 C). These changes were primarily caused by more frequent transitions from shrinkage to pause, and less frequent transitions from pause to shrinkage (Table 1). These differences were not found in cells expressing NAV1-EBmut, demonstrating that NAV1 requires MT plus-end targeting through EB1 to alter MT dynamics (Table 1 and Fig. S5 C). Strikingly, no other transitions in MT dynamics were affected by NAV1 overexpression. Altogether, our findings show that NAV1 does not interfere with MT polymerization but does promote MT plus-end rescues, prevent catastrophes, and induce longer periods of pause in the F-actin–rich cell periphery, which suggests a role of NAV1 in the persistence of MTs in the P-domain of GCs.
Figure 4.
NAV1 promotes the persistence of MTs in the P-domain of GCs. (A) Immunostaining of MTs (α-tubulin) and F-actin (Phalloidin) in axonal GCs of cortical neurons (3 DIV) transduced with the indicated shRNAs by lentiviral infection at 0 DIV. Red lines delineate phalloidin-positive areas. Scale bar, 5 µm. (B) Mean MT fluorescence intensity inside the F-actin–delineated areas of GCs and in the axon shafts of neurons from A. Values are normalized to control. n = 29 (shRNA-luc), 31 (shRNA-NAV1#1,) and 18 (shRNA-NAV1#2) GCs from four independent experiments. (C) EB3-GFP time-lapse projections with comet tracks from axonal GCs of cortical neurons (3 DIV) transfected with EB3-GFP, Lifeact-Tq, and the indicated shRNAs. Dashed lines delineate Lifeact-Tq areas (not shown). Scale bar, 2 µm. (D) Density of EB3-GFP comets inside F-actin. n = 35 (shRNA-luc), 33 (shRNA-NAV1#1), and 29 (shRNA-NAV1#2) GCs from four independent experiments. (E) Density of EB3-mCherry comets inside F-actin in GCs of cortical neurons (2 DIV) transfected with EB3-mCherry, Lifeact-Tq, and the indicated forms of GFP-NAV1, and imaged as in C. n = 42 (GFP), 51 (WT), and 33 (EBmut) GCs from five independent experiments. (F and G) plusTipTracker analysis of the EB3 comet dynamics inside F-actin–rich areas of GCs imaged in D and E, respectively. n = 13 (shRNA-luc), 8 (shRNA-NAV1#1), and 9 (shRNA-NAV1#2) GCs from three independent experiments. n = 22 (GFP), 24 (WT), and 27 (EBmut) GCs from three independent experiments. Histograms show means ± SEM. Analyzed by two-tailed Student's t test.
Figure S5.
Kymograph-based analysis of MT dynamics in LLCPK-1α cells stably expressing GFP-α-tubulin and transfected with mCherry-NAV1. Related to Table 1. (A) Representative TIRF microscopy image of an LLCPK-1α cell showing GFP-labeled MTs exploring a lamellipodium (dashed line). Scale bar, 10 µm. (B) Representative kymographs obtained from time-lapse microscopy of MT plus ends in LLCPK-1α cells stably expressing GFP-α-tubulin and transfected with mCherry-NAV1, illustrating the persistence of mCherry-NAV1 at the tip of paused MTs. Arrowheads point to pausing phases of the MT dynamics. See also Video 6. (C) Representative kymographs depicting the dynamics of MTs in cells transfected with either mCherry (control), mCherry-NAV1-WT, or mCherry-EBmut and imaged by TIRF time-lapse microscopy for 2 min. The ectopic expression of NAV1-WT induces longer periods of pause and shorter catastrophe events resulting from more frequent transitions from shrinkage to pause and less frequent transitions from pause to shrinkage (see Table 1).
Video 6.
mCherry-NAV1 tracks MT plus ends in LLCPK-1α cells. Related to Table 1 and Fig. S5. Time-lapse microscopy of LLCPK-1α cells stably expressing GFP-α-tubulin and transfected with either mCherry-NAV1-WT (left) or mCherry-NAV1-EBmut (right). Time interval: 1 second. Scale bar, 5 µm.
Table 1. Dynamic parameters of MTs measured at the cell cortex of LLCPK-1α cells stably expressing GFP-α-tubulin upon ectopic expression of WT or mutant (EBmut) mCherry-NAV1.
| Dynamic parameters | mCherry | mCh-NAV1-WT | mCh-NAV1-EBmut |
|---|---|---|---|
| Growth | |||
| Duration (s) | 9.37 ± 6.47 | 8.78 ± 5.40 | 8.35 ± 4.92 |
| Rate (μm/min) | 9.32 ± 3.65 | 8.69 ± 4.19 | 9.48 ± 3.96 |
| Shrinkage | |||
| Duration (s) | 2.46 ± 1.93 | 2.02 ± 1.60a | 2.38 ± 1.82b |
| Rate (μm/min) | 42.33 ± 19.09 | 44.59 ± 26.69 | 41.92 ± 24.47 |
| Pause duration (s/min) | 30.50 ± 11.26 | 37.23 ± 13.19c | 36.13 ± 11.85c |
| Transition frequencies | |||
| Growth-shrinkage (min−1) | 3.05 ± 2.11 | 3.50 ± 2.51 | 2.58 ± 2.24 |
| Growth-pause (min−1) | 3.76 ± 2.58 | 4.19 ± 4.04 | 4.92 ± 2.89 |
| Shrinkage-growth (min−1) | 19.02 ± 10.48 | 18.78 ± 12.98 | 20.88 ± 12.34 |
| Shrinkage-pause (min−1) | 6.87 ± 8.12 | 13.29 ± 12.51a | 6.10 ± 9.32b |
| Pause-growth (min−1) | 1.54 ± 1.32 | 1.49 ± 1.28 | 1.29 ± 1.28 |
| Pause-shrinkage (min−1) | 2.27 ± 1.34 | 1.48 ± 1.29a | 2.01 ± 1.72b |
| Number of MTs analyzed | 69 | 65 | 69 |
| Number of cells analyzed | 30 | 27 | 29 |
The average growth and shrinkage durations and rates are the average of all instantaneous durations and rates for the population of MTs. The average pause duration and transition frequencies are the average pause duration and transition frequencies for each MT. Values are means ± SD. Analyzed by two-tailed Student's t test.
Significant compared with mCherry-expressing cells (P < 0.001).
Nonsignificant compared with mCherry-expressing cells (P > 0.05) and significant compared with mCherry-NAV1-WT–expressing cells (P < 0.05).
Significant compared with mCherry-expressing cells (P < 0.01).
NAV1 binds MTs directly and persists at nonpolymerizing MT plus ends
To better understand how NAV1 stabilizes MTs in F-actin–rich domains of GCs, we focused our studies on the dynamics of GFP-NAV1 in the P-domain. We noticed that the forward tip-tracking motion of GFP-NAV1 in filopodia was often followed by a rearward movement toward the central domain (Fig. 5 A and Video 4). Kymograph analysis revealed that this retrograde motion of NAV1 granules was concomitant with the disassembly of the EB complex and hence with a halt in MT growth (Fig. 5, A and B). These EB3-free GFP-NAV1 granules, which were previously reported in GFP-NAV1–expressing cell lines (van Haren et al., 2009), are analogous to the EB1-negative endogenous NAV1 granules that we observed in axonal GCs (Fig. S1 B). Interestingly, some EB3/NAV1-positive comets near the submembranous actin cortex were also found to turn into pausing NAV1 granules upon dissociation of EB3 (Fig. 5 C and Video 7). Our analysis revealed that 72% of the EB3-NAV1–colabeled comets turned into EB-free NAV1 granules, with 81% of those displaying retrograde translocation (RT) and 19% remaining paused during 22.8 ± 8 s (Fig. 5 D). We hypothesized that these EB-negative NAV1 granules could remain in contact with nonpolymerizing MT plus ends. To address this question, we incubated neurons expressing GFP-NAV1 with the fluorescent dye SiR-tubulin, which labels MTs, and monitored both NAV1 and MT dynamics in axonal GCs. We found that ∼75% of the NAV1 granules either retracting or stalling were localized at a MT tip and moved coordinately with it (Fig. 5, E and F; and Video 8). To strengthen these data, we immunostained endogenous NAV1, EB1, and MTs in neurons and imaged GCs using high-resolution microscopy. NAV1 was found colocalizing with EB1 on 57% of the MT tips but, interestingly, NAV1 was also found in the absence of EB1 on 18% of the MT tips present in the P-domain (Fig. 5, G and H). To test whether NAV1 has the ability to bind autonomously to nonpolymerizing MT tips, we performed an in vitro cosedimentation assay with preassembled Taxol-stabilized MTs and a recombinant fragment of NAV1 comprising its MT-binding domain (His-NAV1-N1; Martínez-López et al., 2005). These experiments revealed that NAV1 binds to MTs autonomously, with a calculated Kd of 1.41 ± 0.17 µM (Fig. 5, I and J). These data suggest that, unlike most +TIPs, which bind exclusively growing MT plus ends, NAV1 is able to persist at the plus end of pausing or retracting MTs embedded within the F-actin network of axonal GCs by interacting directly with tubulin.
Figure 5.
NAV1 binds directly and persists at the plus end of nongrowing MTs. (A) Left: Axonal GC of a cortical neuron transfected with GFP-NAV1, EB3-mCherry, and Lifeact-Tq. Right: Time-lapse composition of the EB3-mCherry/Lifeact-Tq and EB3-mCherry/GFP-NAV1 overlays from one filopodium (white box). Closed arrowheads point to a polymerizing EB3/NAV1-positive MT plus end, and open arrowheads point to the NAV1 granular signal sliding backward after the EB3 signal fades up. Scale bars, 2 µm; time, minutes:seconds. See also Videos 4 and 5. (B) Kymographs from the filopodium shown in A. Arrowheads mark the backward motion of the EB3-negative NAV1 granule. (C) Kymographs from the filopodium shown in Video 7. Arrowheads mark a pausing EB3-negative NAV1 granule. (D) Percentage of NAV1/EB3-positive comets turning into EB3-negative NAV1 granules and the proportion of those undergoing retrograde translocation (RT) or pause. The average duration of the NAV1 granule pauses is also detailed. n = 60 total EB3/NAV1-positive comets in 26 GCs from five independent experiments. (E) Time-lapse composition of the axonal GC of a cortical neuron (2 DIV) transfected with GFP-NAV1 and Lifeact-Tq and incubated with SiR-tubulin (MT in red). Arrowheads point to a NAV1 granule at the retracting tip of an MT. F-actin area is defined with dashed lines. n = 56 NAV1 granules tracked in 26 GCs from three independent experiments. Scale bar, 2 µm; time, minutes:seconds. See also Video 8. (F) Kymographs of the MT pointed to in E depicting the coordinated motion of the NAV1 granule (green) and the MT tip (red). (G) Immunostaining of NAV1, EB1, and MTs (α-tubulin) in the axonal GC of a cortical neuron (2 DIV). The dashed square outlines the magnified area. Closed arrowheads point to EB1/NAV1-labeled MT plus ends, and open arrowheads point to MT plus ends that are labeled with NAV1 only. Scale bars, 2 µm. (H) Percentage of MT plus ends at the P-domain in G that are labeled with NAV1 only, EB1/NAV1, EB1 only, or free. n = 80 MT plus ends in 15 axonal GCs. (I) Immunoblots of the soluble (S) and pellet (P) fractions of an in vitro MT cosedimentation assay with purified His-NAV1-N1 fragment. Results were replicated three times. (J) Saturation binding curve for His-NAV1-N1 and the equilibrium dissociation constant (Kd) calculated as described in Materials and methods.
Video 7.
GFP-NAV1 tracking a polymerizing MT plus end inside a filopodium turns into a pausing GFP-NAV1 granule devoid of EB3-mCherry. Related to Fig. 5 C. Time-lapse microscopy of a GC filopodium from a neuron transfected with GFP-NAV1, EB3-mCherry, and Lifeact-Tq. Overlays of Lifeact/EB3 (top) and NAV1/EB3 (bottom) are shown. From the 45th second of the movie on, the EB3 signal disappears while the NAV1 granule signal persists. Time interval: 1 second.
Video 8.
GFP-NAV1 persists at the tip of nonpolymerizing MTs within the F-actin–rich domain of GCs. Related to Fig. 5 E. Time-lapse microscopy of the axonal GC of a neuron transfected with GFP-NAV1 and Lifeact-Tq, with MTs labeled by SiR-tubulin (red). Overlays of NAV1/MTs (left) and NAV1/MTs/F-actin (right) are shown. The F-actin network is pulled backward by the translocating NAV1-labeled MTs. Time interval: 1 second.
NAV1 crosslinks nonpolymerizing MT plus ends to actin filaments in filopodia
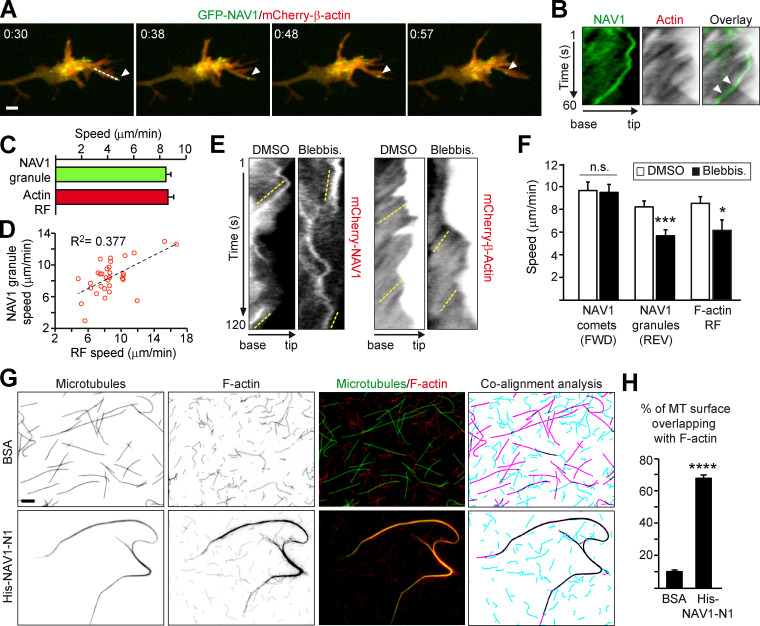
The retrograde motion of GFP-NAV1 granules observed in the P-domain of GCs was reminiscent of the F-actin retrograde flow (RF; Forscher and Smith, 1988). Since NAV1 binds F-actin, we conceived that NAV1 granules may link nonpolymerizing MT plus ends to actin fibers, thereby causing a backward movement of MT tips coordinately with the RF. To address this hypothesis, we compared the velocity of NAV1 granules and the RF in filopodia of axonal GCs of neurons coexpressing GFP-NAV1 and mCherry-β-actin (Fig. 6, A and B). Interestingly, kymograph analysis revealed similar average speeds for NAV1 granules and for the RF and a significant correlation between NAV1 granules and RF speeds in each filopodium analyzed (Fig. 6, C and D). Of note, our average RF speed values were similar to those previously reported under the same experimental conditions (Geraldo et al., 2008; Neukirchen and Bradke, 2011). To further support a functional link between NAV1-labeled nonpolymerizing MTs and the RF, we used the myosin II–specific inhibitor blebbistatin to slow down the RF and examined the effect on the dynamics of NAV1 granules. Neurons expressing either mCherry-β-actin or mCherry-NAV1 were exposed to blebbistatin or vehicle, and we measured the speed of the RF, NAV1 comets, and NAV1 granules in GC filopodia (Fig. 6 E). Dual imaging of actin and NAV1 using a GFP reporter was not possible due to blebbistatin-induced phototoxicity when it is excited with blue light (Kolega, 2004). Blebbistatin reduced the speed of NAV1 granules moving backward to a similar extend as the RF (Fig. 6, E and F). In contrast, it did not affect the speed of NAV1 comets moving forward, demonstrating the absence of effect on the polymerization of NAV1-decorated MTs. These results show that the retrograde translocation of nongrowing MT tips carrying NAV1 is influenced by the RF in GC filopodia, suggesting that NAV1 may crosslink nongrowing MT plus ends to actin filaments. To obtain direct evidence of a MT–F-actin crosslinkage via NAV1, we performed TIRF microscopy with fluorescently labeled stabilized MTs and actin filaments incubated with recombinant His-NAV1-N1, which comprises both MT-binding and actin-binding domains of NAV1. In the control condition, coincubated MTs and actin filaments remained well individualized. By contrast, the addition of His-NAV1-N1 induced a remarkable coalignment of actin filaments with MT bundles, with 68% of the surface of MTs overlapping with actin filaments in the presence of His-NAV1-N1 compared with only 10% in the presence of BSA (Fig. 6, G and H). The ability of NAV1 to simultaneously bind and crosslink the two cytoskeletal systems in vitro, together with our previous findings, strongly suggests that NAV1 is involved in the coordination of the actin and MT cytoskeleton in GCs.
Figure 6.
NAV1 crosslinks nonpolymerizing MT plus ends to actin filaments in filopodia. (A) Time-lapse composition of the axonal GC of a cortical neuron transfected with GFP-NAV1 and mCherry-β-actin. Arrowheads mark a NAV1 granule inside a filopodium. Scale bar, 2 µm. time, minutes:seconds. (B) Kymographs generated from A (dashed line) to measure the speed of NAV1 granules and the F-actin RF. Arrowheads point to a granule aligned with the RF lines. (C) Average speed of NAV1 granules and the RF from B. n = 32 GFP-NAV1 granules in 20 GCs from three independent experiments. (D) Correlation between NAV1 granules and RF speeds in each filopodium from B. P < 0.01 in a Spearman correlation test. (E) Kymographs obtained from the filopodia of axonal GCs of cortical neurons (2 DIV) transfected with either mCherry-NAV1 or mCherry-β-actin and treated with blebbistatin or DMSO. Dashed yellow lines mark the retrograde motion of NAV1 granules and the RF. (F) Average speed of the NAV1 comets (FWD) and granules (REV) measured in kymographs from E. n = 41 comets and 45 granules in 26 GCs (DMSO), and 38 comets and 42 granules in 28 GCs (blebbistatin), from three independent experiments. Average speed of the RF measured in kymographs from E. n = 21 (DMSO) and 13 (blebbistatin) filopodia in 18 and 15 GCs, respectively, from two independent experiments. (G) Fluorescently labeled Taxol-stabilized MTs (green) and phalloidin-stabilized actin filaments (red) incubated with 0.2 µM His-NAV1-N1 fragment or BSA as a negative control. The masked images (right panel) represent MTs (magenta), actin filaments (turquoise), and their overlapping areas (black). Scale bar, 5 µm. (H) Quantification of the percentage of MT surface overlapping with F-actin from G. n = 28 (BSA) and 25 (His-NAV1-N1) images from two independent experiments. Histograms show means ± SEM. Analyzed by two-tailed Student's t test.
NAV1 participates in the radial migration of cortical projection neurons during corticogenesis
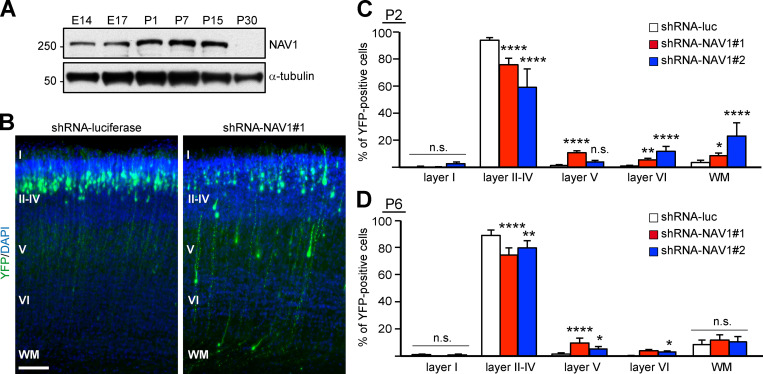
NAV1 is expressed in migratory neurons in the cerebral cortex and cerebellum during embryonic development (Martínez-López et al., 2005). We found that NAV1 is expressed in mouse cortical extracts obtained from embryonic day 14 (E14) to postnatal day 15 (P15), but its level declines thereafter and is barely detectable at P30 (Fig. 7 A). To uncover a potential role of NAV1 in the migration of cortical neurons in vivo, we depleted NAV1 during embryogenesis by using shRNAs. Briefly, plasmids coexpressing control or NAV1-targeting shRNAs and a YFP reporter were transfected into radial glia progenitors at the dorsolateral ventricular zone of the mouse forebrain at E15.5 by in utero electroporation, and the radial migration of projection neurons was analyzed at P2. Consistent with previous reports (Wang et al., 2007), most of the YFP-expressing control neurons had completed their migration into cortical layers II–IV by P2, and only a residual proportion appeared in the subgranular layers (V, VI, and white matter; Fig. 7, B and C). By contrast, we measured a decreased proportion of NAV1-deficient neurons reaching layers II–IV concomitant with an increased proportion of neurons remaining in deeper layers (Fig. 7, B and C). Given that the radial migration of the cortical neurons is completed by P7 in mice (Jeanneteau et al., 2010; Wang et al., 2007), we analyzed whether the migration delay derived from NAV1 depletion persisted at this stage. Similar to the results obtained at P2, NAV1-depleted neurons had not reached their final destination in the upper cortical layers at P6 (Fig. 7 D). Therefore, our data demonstrate that NAV1 plays a role in the radial migration and final layer positioning of cortical projection neurons during embryonic development.
Figure 7.
NAV1 participates in the radial migration of cortical projection neurons in the developing cerebral cortex. (A) Immunoblots of lysates of brain cortex at the indicated embryonic (E) and postnatal (P) days. α-Tubulin was used as a loading control. Results were replicated twice. (B) Coronal sections through the somatosensory cortex of P2 pups electroporated with the indicated shRNAs. DAPI was used for identification of cortical layers, labeled with roman numbers. WM, white matter. Scale bar, 100 µm. (C) Distribution of YFP-expressing cells in the indicated cortical layers from B. n = 4 (shRNA-luc), 3 (shRNA-NAV1#1), and 2 (shRNA-NAV1#2) mice. (D) Distribution of YFP-expressing cells in sections of P6 pups as in B. n = 4 mice per condition. Histograms show means ± SEM. Analyzed by χ2 test.
Discussion
The transient coupling of MTs to the actin cytoskeleton has emerged as a key molecular mechanism to carry out morphogenic and migratory processes of neurons during brain development (Cammarata et al., 2016; Geraldo and Gordon-Weeks, 2009). In this study, we unravel new functions of the +TIP NAV1 in promoting F-actin–MT crosslinking and stabilizing MT plus ends in the F-actin–rich P-domain of axonal GCs, with physiological implications in GC dynamics and neuronal motility.
We report that NAV1 is a +TIP with autonomous F-actin–binding properties, which may account for its accumulation in F-actin–rich domains at the periphery of GCs. NAV1 binds F-actin through an N-terminal region that is not structurally related to any of the known conserved families of domains found in actin-binding proteins, such as CH domains contained by ACF7, or dis1/TOG domains contained by CLASP and XMAP215 (Leung et al., 1999; Slater et al., 2019; Tsvetkov et al., 2007). Our study also provides new molecular insights into how NAV1 binds to MTs. We show that the recruitment of NAV1 to MT plus ends relies on its interaction with EB1 via two SxIP motifs located in the N-terminus of NAV1. We also find that unlike most EB-dependent +TIPs, NAV1 persists in the form of granules at the plus end of nongrowing MTs after disassembly of the EB complex and binds directly to MTs in vitro through its N-terminal part comprising the predicted MT-binding domain. A similar EB-independent association to MT tips has been reported notably for XMAP215, which binds MTs autonomously via its TOG domains, and the Drosophila orthologue of ACF7, which binds the MT lattice via a growth arrest specific 2 domain (Brouhard et al., 2008; Alves-Silva et al., 2012). The N-terminal region of NAV1 does not contain any TOG nor growth arrest specific 2 domains, so further structure–function analyses will be required to define more precisely the MT-binding site of NAV1.
By combining both loss- and gain-of-function approaches with cultured cortical neurons, we show that NAV1 controls GC morphology and steering. GCs of NAV1-depleted neurons are enlarged, resembling the phenotypes described upon knockdown of the F-actin–MT crosslinkers CLIP170, APC, XMAP215, and Fidgetin-like1 protein, which is a member of the MT-severing protein family (Fassier et al., 2018; Neukirchen and Bradke, 2011; Purro et al., 2008; Slater et al., 2019). In addition, our data reveal that the enlargement of GCs caused by NAV1 depletion is accompanied by a reduction in the density of MTs in the P-domain of GCs, similarly to what was observed upon depletion of the F-actin–MT crosslinker CLIP170 (Neukirchen and Bradke, 2011). This suggests that NAV1 stabilizes MTs in F-actin–rich regions of the GCs, but we cannot rule out the possibility that NAV1 may also facilitate the MT invasion into the F-actin network. Consistent with the former hypothesis, we found that NAV1 prevents the depolymerization of MTs at the F-actin–MT interface and extends their overall pause duration in nonneuronal cells, most likely accounting for longer MT persistence in F-actin–rich areas of GCs. This property requires NAV1 operating at the tip of nongrowing MTs, which is in agreement with our demonstration that NAV1 can bind MTs independently of EB1. NAV1 requires binding to EB1 and tracking MT plus ends to both stabilize MTs and rescue the defects of NAV1-depleted GCs. This strongly suggests that NAV1 is primarily recruited to the tip of growing MTs by EB1, and then interacts with tubulin directly, which enables it to persist at MT tips independently of EB1. It is possible that the interaction of NAV1 with EB1 at the plus end of MTs changes its conformation, thus opening domains for MT attachment, similarly to what was proposed for Shot, the Drosophila orthologue of ACF7 (Applewhite et al., 2013). In addition, the GC phenotypes induced by NAV1 manipulation may also reflect a dysregulation of F-actin remodeling. Consistent with this, previous studies have shown that NAV1 can influence F-actin dynamics (Marcus-Gueret et al., 2012; Pandey et al., 2018; Schmidt et al., 2009; van Haren et al., 2014) and, therefore, future work should seek to address the contribution of NAV1 to F-actin remodeling.
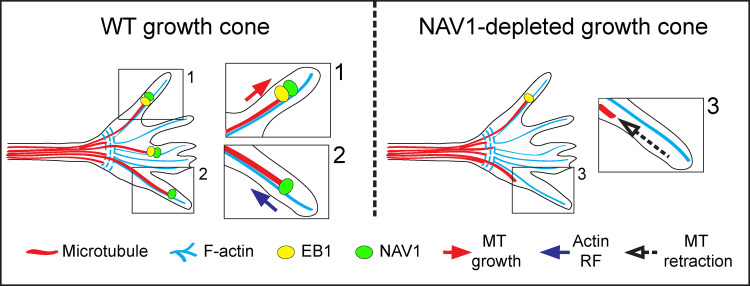
In addition to the MT-stabilizing function of NAV1, we found that (1) NAV1-labeled MT plus ends can slide backward synchronously with the F-actin RF in GCs; and (2) NAV1 binds F-actin and MTs simultaneously in vitro, mediating their coalignment. These data support an F-actin–MT crosslinking activity of NAV1 in GCs. Some actin-binding +TIPs, such as EB3, XMAP215, and APC, mediate transient coupling events that promote F-actin–MT coalignment to drive MT polymerization along preexisting F-actin bundles (Geraldo et al., 2008; Slater et al., 2019; Zhou et al., 2004). CLASP2 instead promotes a sustained crosslinking between MT plus ends and F-actin, causing MT looping and growth attenuation inside GCs due to the RF (Hur et al., 2011; Lee et al., 2004). In contrast, overexpression of NAV1 does not induce MT looping or alter the MT polymerization rate in GCs. Furthermore, slowing down the F-actin RF by blebbistatin treatment does not accelerate the polymerization of NAV1-decorated MT plus ends in the neuronal GCs, but it results in a slowdown of the retrograde translocation of NAV1-labeled nonpolymerizing MT tips. Altogether, these data support a model in which NAV1 couples polymerizing MT tips with actin filaments in an EB-dependent fashion that does not alter their polymerization rate. Then, upon dissociation of the EB complex, NAV1 switches to an EB-independent interaction to MT plus ends that enables it to promote their stabilization. The stabilization of nongrowing MT plus ends and their transient crosslink to F-actin by NAV1 may prevent their stochastic exit from the F-actin–rich area of the GC and consequently affect GC dynamics (see model in Fig. 8).
Figure 8.
Proposed model depicting how NAV1 affects GC dynamics. (1) In WT GCs, F-actin-bound NAV1 is recruited by EB1 on polymerizing MTs invading the GC periphery. (2) Once the EB complex disassembles and the MT stops growing, NAV1 remains at the nonpolymerizing MT plus ends by binding MTs directly. NAV1 bridges F-actin and nongrowing MTs, which either pause or are subjected to the F-actin RF. This NAV1-mediated crosslink prevents the exit of MT plus ends from the actin network. (3) NAV1 depletion reduces the number of nonpolymerizing MTs crosslinked to F-actin, resulting in their premature exit from the F-actin–rich area. We hypothesize that the ability of NAV1 to promote the persistence of MTs and their asymmetric capture by the F-actin network in the GC periphery is determinant to control GC dynamics and steering in response to guidance cues.
Axonal GCs of NAV1-depleted neurons progress in vitro with more frequent spontaneous changes of direction than control neurons, and they are unresponsive to netrin-1. It is known that the invasion and selective stabilization of MTs by +TIPs into the periphery of the GC are determinant for axon steering (Koester et al., 2007). Complementarily, molecular motors such as dynein or kinesin-5 play a prominent role in the redistribution of MTs during GC turning (Kahn and Baas, 2016). In light of our results, we hypothesize that NAV1 may control the axial steering of the GC by promoting the asymmetric capture of nonpolymerizing MTs by the peripheral F-actin network in response to netrin-1 and/or by controlling the exposure of the netrin-1 receptor Deleted in Colorectal Cancer (DCC) at the surface of GCs via MT-dependent transport.
Previous studies showed that UNC-53 plays a role in cellular migratory movements and that NAV1 mediates the attraction of migrating hindbrain neurons toward a gradient of netrin-1 in vitro (Martínez-López et al., 2005; Stringham et al., 2002). In line with these findings, our work assigns to NAV1 a role in the radial migration of neurons through the developing mammalian neocortex similar to other F-actin–binding +TIPs, such as ACF7 and CLASP2 (Dillon et al., 2017; Ka et al., 2014). Depleting the netrin-1 receptor DCC leads to a delay in cortical radial migration due to defects in the multipolar-to-bipolar transition of the migrating neurons (Zhang et al., 2018). Therefore, the migration delay elicited by NAV1 depletion may be assigned to an impaired netrin-1–DCC signaling in the migrating cortical neurons.
In summary, here we describe a new mechanism of reversible coupling between the MT and actin networks mediated by the +TIP NAV1 in the axonal GC of cortical neurons. We provide evidence that this mechanism regulates MT dynamics and contributes to the control of GC dynamics and motility. Our study emphasizes the functional diversity of the components involved in the F-actin–MT interplay, which is an essential process not only for neuronal connectivity but also for a wide range of cellular processes.
Materials and methods
Animals, cell culture, and transfection
Pregnant female mice (Mus musculus; strain RjOrl:SWISS) were purchased from Janvier Labs and maintained at the BioCampus animal facility (Réseau des animaleries de Montpellier, Montpellier, France), in strict accordance with the French and European Union regulations. Work protocols have been approved by the Local Care and Use Committee (agreement number F34-172-16), in accordance with applicable legislation. Cortical cell cultures were prepared from E14.5 mouse embryos (Sánchez-Huertas et al., 2016). Briefly, cortical tissue was dissected, treated with 0.25% trypsin (Life Technologies) for 15 min at 37°C, and dissociated into single cells by gentle trituration. Neurons were seeded on glass coverslips, glass-bottom plates, or plastic plates coated with 0.1 mg/ml poly-L-lysine (Sigma-Aldrich) at ∼105 cells/cm2 or 2 × 104 cells/cm2 for low-density cultures. Neurons were plated in DMEM containing 10% (vol/vol) FBS, 50 U/ml penicillin, and 50 µg/ml streptomycin (pen/strep), and 1–2 h later the medium was replaced with Neurobasal medium supplemented with 2% B27, pen/strep, 0.6% glucose and 1% Glutamax (all reagents from Life Technologies). Low-density cultures were supplemented at day-in-vitro (DIV) 1 with conditioned media from mature cultures. HEK293T and HeLa cells were grown in DMEM containing 10% (vol/vol) FBS, 2 mM L-glutamine, and pen/strep. LLCPK-1α cells stably expressing GFP-α-tubulin were a gift from P. Wadsworth (Rusan et al., 2001) to B. Delaval and were cultured in medium consisting of 45% Opti-MEM Reduced Serum Medium, 45% Ham's F-10 Nutrient Mix, 10% (vol/vol) FBS, and 2 mM L-glutamine. All cells were kept at 37°C in a humidified atmosphere containing 5% CO2. Cultured cortical neurons were transfected at 1 DIV using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Transfection of cDNA constructs into HEK293T and HeLa cells was performed with JetPEI and into LLCPK-1α with JetPRIME (both from Polyplus Transfection) according to the manufacturer's instructions. Transfection of siRNAs into HeLa cells was performed with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions.
Expression constructs
The pbio-EGFP-NAV1 construct has been described previously (van Haren et al., 2009). To generate pmCherry-NAV1, an EcoRV(blunted)/MfeI fragment from pbio-EGFP-NAV1 was subcloned into the XhoI(blunted)/MfeI sites of the pmCherry-C1 vector. The pbio-EGFP-NAV1 single (SRIP/SRNN and SGIP/SGNN) and double (NAV1-EBmut) point mutants were generated by PCR-based site-directed mutagenesis using the CloneAmp HiFi PCR premix (Clontech). pmCherry-NAV1-EBmut was generated by replacing the XmnI/PshAI fragment of pmCherry-NAV1 with the XmnI/PshAI fragment of pbio-EGFP-NAV1-EBmut encompassing both SxIP mutations. To construct pbio-EGFP-NAV1-N1 (aa 1–762), pbio-EGFP-NAV1-C1 (aa 763–1836), and pbio-EGFP-NAV1-N5 (aa 424–762), the specified NAV1 regions in pbio-EGFP-NAV1 were PCR amplified and inserted into the BspEI site of the pbio-EGFP-C1 vector using the In-Fusion HD Cloning kit (Clontech). To construct pbio-EGFP-NAV1-N2 (aa 1–423), an Eco47III/SpeI(blunted) fragment was generated from pbio-EGFP-NAV1 and religated. To construct pbio-EGFP-NAV1-C2 (aa 1047–1836), a BamHI/BglII fragment was generated from pbio-EGFP-NAV1 and religated. To construct pbio-EGFP-NAV1-N3 (aa 214–762), the specified NAV1 region in pbio-EGFP-NAV1 was PCR amplified and ligated into the BglII/SalI sites of the pbio-EGFP-C1 vector. To construct pbio-EGFP-NAV1-N4 (aa 213–516), the specified NAV1 region in pbio-EGFP-NAV1 was PCR amplified and ligated into the BspEI/SalI sites of the pbio-EGFP-C1 vector. To generate the constructs encoding His-NAV1-N1 (aa 1–762) and His-NAV1-N5 (aa 424–762), the specified NAV1 regions in pbio-EGFP-NAV1 were PCR amplified and inserted into the NheI site of the pET-28a vector using the In-Fusion HD Cloning kit. To construct His-NAV1-N6 (aa 604–762), the specified NAV1 region in pbio-EGFP-NAV1 was PCR amplified and ligated into the NdeI/SalI sites of the pET-28a vector. The pSUPER-mCherry-Neo plasmids encoding NAV1-targeting and luciferase shRNAs have been described previously (van Haren et al., 2014). For lentivirus production, the target sequences to deplete NAV1 (NAV1#1: 5′-CCTAGAACAGCTACTCTTC-3′; NAV1#2: 5′-CCATGATGCAGCACTACAT-3′) were cloned for expression as shRNAs into the pLL3.7 vector encoding GFP as a reporter in a separate reading frame (plasmid #11795; Addgene; (Rubinson et al., 2003). To generate these constructs, sense and antisense oligonucleotides were annealed and ligated into the HpaI/XhoI sites of the pLL3.7 vector. The pLL3.7 plasmid encoding luciferase shRNA (5′-CTTACGCTGAGTACTTCGA-3′) was kindly provided by J. Lüders. For in utero electroporations, the shRNA expression cassettes were PCR amplified from the pLL3.7 constructs and ligated into the KpnI site of the pAAV-CAG-YFP vector (plasmid #104055; Addgene; (Challis et al., 2019). All the primers used for cloning are listed in Table 2. Constructs encoding GST-EB1 and EB3-mCherry have been described before (Stepanova et al., 2003). The pLifeact-mTurquoise plasmid was kindly provided by J. Goedhart. The pEGFP-β-actin and pmCherry-β-actin plasmids were gifts from S. Bodin. The siRNA duplexes targeting EB1 and the non-targeting control siRNA#1 were synthesized by Horizon Discovery. Sequences used were as follows: EB1#1, 5′-UUCGUUCAGUGGUUCAAGAdTdT-3′ (Watson and Stephens, 2006); EB1#2, 5′-UUGCCUUGAAGAAAGUGAA-3′ (Louie et al., 2004).
Table 2. Oligonucleotides used in this study.
| Construct | Oligonucleotide (5′–3′) |
|---|---|
| GFP-NAV1-SRIP/SRNN | CCTGGGAAAGCCGAGCCGGAACAATCGAGGACCCTATGCGGAGGTC |
| GACCTCCGCATAGGGTCCTCGATTGTTCCGGCTCGGCTTTCCCAGG | |
| GFP-NAV1-SGIP/SGNN | GATCCAGAAGTCCTCAGGCAACAATGTCAAGCCAGTAAATGGGCG |
| CGCCCATTTACTGGCTTGACATTGTTGCCTGAGGACTTCTGGATC | |
| GFP-NAV1-N1 | GCTGTACAAGTCCGGACTGCCCAAGCGCGCCAAGG |
| GAGATCTGAGTCCGGATTACTTGTCAAGATTGGCTAGTGAGG | |
| GFP-NAV1-N3 | TCTTAGATCTCATATGGAGTCCCAGAGAAAGAGG |
| TCTTGTCGACCTACTTGTCAAGATTGGCTAG | |
| GFP-NAV1-N4 | GGTTCCGGACCAGAGTCCCAGAGAAAG |
| AGGTGTCGACTTAAGGTTTGCCTGCGACTTT | |
| GFP-NAV1-N5 | GCTGTACAAGTCCGGACTGGCAGAAAGTGGGCTGAG |
| GAGATCTGAGTCCGGATTACTTGTCAAGATTGGCTAGTGAGG | |
| GFP-NAV1-C1 | GCTGTACAAGTCCGGAGTCAACTCCAACAGTCTGGATC |
| GAGATCTGAGTCCGGATTAAAGTGTTGCCTGAAGGTTGGG | |
| His-NAV1-N1 | CAGCCATATGGCTAGCCTGCCCAAGCGCGCCAAGG |
| CACCAGTCATGCTAGCTTACTTGTCAAGATTGGCTAGTGAGG | |
| His-NAV1-N5 | CAGCCATATGGCTAGCCTGGCAGAAAGTGGGCTGAG |
| CACCAGTCATGCTAGCTTACTTGTCAAGATTGGCTAGTGAGG | |
| His-NAV1-N6 | TCTTAGATCTCATATGCGCAAGACTAGCTTAGATGT |
| TCTTGTCGACCTACTTGTCAAGATTGGCTAG | |
| shRNA-NAV1#1 (pLL3.7) | TGCCTAGAACAGCTACTCTTCTTCAAGAGAGAAGAGTAGCTGTTCTAGGCTTTTTTC |
| TCGAGAAAAAAGCCTAGAACAGCTACTCTTCTCTCTTGAAGAAGAGTAGCTGTTCTAGGCA | |
| shRNA-NAV1#2 (pLL3.7) | TGCCATGATGCAGCACTACATTTCAAGAGAATGTAGTGCTGCATCATGGCTTTTTTC |
| TCGAGAAAAAAGCCATGATGCAGCACTACATTCTCTTGAAATGTAGTGCTGCATCATGGCA | |
| shRNAs (pAAV-CAG-YFP) | TTGGTACCCAGCAGAGATCCAGTTTGG |
| TTGGTACCGAAGCGAGCTTATCGATACC |
Antibodies and drugs
The mouse antibodies used were anti-NAV1 (sc-398641; Santa Cruz Biotechnology; dilution for immunofluorescence [IF]: 1:250, for Western blotting [WB]: 1:1,000), anti-EB1 (610534; BD Biosciences; dilution IF: 1:200), anti-α-tubulin (T6199; Sigma-Aldrich; dilution IF: 1:4,000, dilution WB: 1:10,000), anti-6xHis (552565; BD Biosciences; dilution WB: 1:5,000), and anti-actin (clone C4; MP Biomedicals; dilution WB: 1:10,000). The chicken anti-GFP (GFP-1010; Aves Labs; dilution IF: 1:1,000), the rabbit anti-GFP (TP401; Torrey Pines Biolabs; dilution WB: 1:10,000), and the rat anti-α-tubulin (clone YL1/2; dilution IF: 1:1,000), kindly provided by K. Rogowski (Institut de Génétique Humaine, Montpellier, France), were also used. DyLight Fluor-conjugated secondary antibodies were from Thermo Fisher Scientific, and horseradish peroxidase-conjugated secondary antibodies were form GE Healthcare. ATTO-488–labeled tubulin was kindly provided by D. Portran. Cytochalasin D and blebbistatin (both from Sigma-Aldrich) were added to the cell cultures 20 min before analysis at 1 µM and 40 µM, respectively. SiR-tubulin (Tebu-Bio) was added to cell cultures 90 min before analysis at 50 nM.
Recombinant protein production
The pET28a constructs encoding 6xHis-tagged fragments of NAV1 and the pGEX-3X constructs encoding GST and GST-EB1 were transformed into Escherichia coli Rosetta(DE3) pLysS and BL21(DE3) competent cells, respectively, and bacteria were grown at 37°C in Luria-Bertani (LB) medium supplemented with 50 µg/ml kanamycin (pET28a) or 100 µg/ml ampicillin (pGEX-3X) to an OD600 of 0.8–1.0. Expression was induced by adding 1 mM IPTG and incubating the bacteria for 2 h at 37°C. Cell pellets were resuspended in a buffer containing 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1% [wt/vol] Triton X-100, 0.2 mg/ml lysozyme, 1 mM DTT, 1 mM benzamidine, and 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride and lysed on ice by ultrasonication. The lysates were clarified by centrifugation at 20,000 g for 20 min at 4°C. For affinity purification of 6xHis-tagged fragments of NAV1, the lysate supernatants were incubated with HIS-Select Nickel Affinity Gel beads (Sigma-Aldrich) for 2 h at 4°C. The beads were washed four times with high-salt buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 0.02% [wt/vol] Triton X-100, and 1 mM DTT), twice with low-imidazole buffer (50 mM Tris-HCl, pH 8.0, 100 mM imidazole, 0.02% [wt/vol] Triton X-100, and 1 mM DTT), and twice with no-salt buffer (50 mM Tris-HCl, pH 8.0, and 1 mM DTT). The proteins were eluted by incubating the beads with a solution of 50 mM Tris-HCl, pH 8.0, 500 mM imidazole, 1 M NaCl, and 1 mM DTT for 30 min at 4°C. The eluted His-NAV1-N1 protein fragment used for in vitro MT cosedimentation assays and in vitro F-actin–MT crosslinking assays was then dialyzed overnight at 4°C in BRB80 buffer (80 mM Pipes, 1 mM EGTA, and 1 mM MgCl2, pH 6.8) containing 10% sucrose and 1 mM DTT. The His-NAV1-N5 and His-NAV1-N6 protein fragments used for in vitro F-actin cosedimentation assays were dialyzed overnight at 4°C in PBS containing 10% sucrose and 1 mM DTT. To generate GST and GST-EB1 affinity columns, glutathione-Sepharose 4B beads (GE Healthcare) were employed. After incubation with the lysates, the beads were washed four times with high-salt buffer, twice with no-salt buffer, and once with no-salt buffer supplemented with 10% sucrose. The resulting affinity columns were aliquoted and stored at −80°C.
GST-EB1 pull-down assays
Lysates of HEK293T cells (50 µg) expressing the indicated GFP-tagged forms of NAV1 were incubated with 2.5 µg GST or GST-EB1 conjugated to glutathione-Sepharose 4B. After incubation for 1 h on a rotating wheel at 4°C, the beads were washed twice with lysis buffer supplemented with 150 mM NaCl, and once with 50 mM Tris-HCl (pH 7.5). The proteins retained on the beads were resuspended in Laemmli buffer and analyzed by Western immunoblotting.
F-actin cosedimentation assays
F-actin cosedimentation assays were performed by using the Actin Binding Protein Biochem Kit (Cytoskeleton) with rabbit skeletal muscle actin, essentially as described by the manufacturer. Briefly, preparations of His-tagged proteins were centrifuged at 150,000 g for 1 h at 4°C to remove aggregates of insoluble material. The supernatants were incubated with 16 µM freshly polymerized F-actin prepared from 20 µM G-actin for 30 min at room temperature. The solutions were then subjected to centrifugation at 150,000 g for 1.5 h at room temperature to pellet actin filaments with bound protein. BSA and α-actinin were used as a negative and positive control, respectively. After solubilization of the pellet fraction in a volume of H2O equal to the initial incubation volume, equal volumes of the pellet and supernatant fractions were resolved by SDS-PAGE. Proteins were then either visualized by Coomassie Blue staining or detected by immunoblotting with the anti-6xHis and anti-actin antibodies. To calculate the equilibrium dissociation constant (Kd) in Fig. S4 D, the fractions of His-NAV1-N6 that cosedimented with F-actin were quantified by scanning densitometry. After subtraction of the amount of His-NAV1-N6 sedimenting in the absence of F-actin, the amount of His-NAV1-N6 cosedimenting with F-actin was converted to moles of His-NAV1-N6 per mole of F-actin and plotted against concentrations of His-NAV1-N6 to derive the maximal molar binding ratio (Bmax) and the Kd.
MT cosedimentation assays
MT cosedimentation assays were performed using the Microtubule Binding Protein Spin-down Assay Kit (Cytoskeleton) with tubulin of porcine brain source, essentially as described by the manufacturer. Briefly, preparations of His-NAV1-N1 were centrifuged at 100,000 g for 30 min at 4°C to remove aggregates of insoluble material. The supernatants were incubated with 2 µM Taxol-stabilized MTs prepared from 50 µM tubulin for 30 min at room temperature. The solutions were then subjected to centrifugation at 100,000 g for 30 min at room temperature to pellet MTs with bound protein. After solubilization of the pellet fraction in a volume of H2O equal to the initial incubation volume, equal volumes of the pellet and supernatant fractions were resolved by SDS-PAGE and immunoblotted with the anti-6xHis and anti-α-tubulin antibodies. The fractions of His-NAV1-N1 that cosedimented with MTs were quantified using LI-COR Odyssey fluorescent imaging and associated Image Studio analysis software. After subtraction of the amount of His-NAV1-N1 sedimenting in the absence of MTs, the amount of His-NAV1-N1 cosedimenting with MTs was converted to moles of His-NAV1-N1 per mole of tubulin and plotted against concentrations of His-NAV1-N1 to derive the equilibrium dissociation constant (Kd).
F-actin–MT crosslinking assays
Fluorescent Taxol-stabilized MTs were prepared from 50 µM tubulin containing 17% of ATTO-488–labeled tubulin. F-actin was prepared from 20 µM G-actin and incubated for 2 min with 2 µM phalloidin-TRITC. Fluorescent Taxol-stabilized MTs (1 µM) and phalloidin-stabilized F-actin (0.5 µM) were incubated with 0.2 µM recombinant His-NAV1-N1 or BSA in BRB80 buffer for 20 min at room temperature. The solutions were diluted 20-fold in BRB80 buffer containing 100 mM DTT and applied to glass coverslips coated with 0.1 mg/ml poly-L-lysine, which were mounted onto glass slides for visualization by fluorescence microscopy.
Cell lysis and Western immunoblotting
Cell cultures and brain cortices were homogenized in ice-cold lysis buffer (50 mM Tris-HCl, pH 7.5, 1 mM EGTA, 1 mM EDTA, 0.1% [wt/vol] Triton X-100, 1 mM sodium orthovanadate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 10% sucrose, 1 mM DTT, 1 mM dithiothreitol, and 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride). Brain tissue was further homogenized using a Potter-Elvehjem homogenizer, and lysates were clarified by centrifugation at 20,000 g for 10 min at 4°C. Protein samples in Laemmli buffer were subjected to SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked for 30 min in TBS-Tween buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, and 0.5% [wt/vol] Tween-20) containing 3% BSA and then incubated overnight with primary antibodies in the same buffer. The blots were washed with TBS-Tween buffer and incubated for 1 h with the appropriate HRP-conjugated secondary antibodies in TBS-Tween buffer containing 3% BSA. After repeating the washing steps, detection was performed using the enhanced chemiluminescence reagent (Millipore). Alternatively, when signal quantification was performed, the immunoblots were incubated with DyLight Fluor-conjugated secondary antibodies (680 or 800 conjugate) and quantified using LI-COR Odyssey fluorescent imaging and associated Image Studio analysis software.
Lentivirus production and transduction
Lentiviruses were generated using the LentiLox3.7 system (Rubinson et al., 2003)at the Vectorology facility of BioCampus (Plateforme de Vectorologie de Montpellier, Montpellier, France). Briefly, HEK293T cells were cotransfected with the pLL3.7 constructs and the packaging plasmids. Lentiviral particles in the medium were concentrated, resuspended in PBS, aliquoted, and stored at −80°C. Infectivity was assayed for GFP-carrying viruses by infecting HEK293T cells with serial dilutions of concentrated lentiviruses and sorting of GFP-positive cells by FACS 72 h after infection. Neurons were infected at 0 DIV at a multiplicity of infection of 4. In the case of low-density cultures, a multiplicity of infection of 7 was used. The complete medium was replaced with fresh medium 16–18 h after infection. The infection efficiencies in cultured neurons were 82% ± 2% and 84% ± 2% for shRNA-luciferase and shRNA-NAV1#1, respectively.
Dunn chamber turning assay
Cortical neurons were grown at low density on square 18 × 18 mm coverslips coated with 0.1 mg/ml poly-L-lysine. Cultures were infected with lentiviruses at 0 DIV, and at 2 DIV, coverslips were assembled in a Dunn chamber (Hawksley). Briefly, coverslips with the infected neurons were inverted over the Dunn chamber leaving a narrow slit on one side, and the three other sides were sealed using a hot paraffin/Vaseline (1:1) mixture. The outer well was refilled carefully with conditioned media containing netrin-1 (R&D Systems) at 200 ng/ml or vehicle, and the narrow slit was sealed (Hutchins et al., 2011; Dudanova et al., 2010; Antoine-Bertrand et al., 2016; DeGeer et al., 2015). Time-lapse phase-contrast images were acquired every 4 min for 2 h at the bridge region of the Dunn chamber with an inverted microscope (IX83; Olympus) using a 10×/0.3 NA PH1 objective and a camera scMOS ZYLA 4.2 MP. Dunn chambers were kept at 37°C in an atmosphere containing 5% CO2 during imaging. Only single axons showing a net growth >9 µm during the 2 h, not touching debris, another cell, or their own cell body, were analyzed. The angle turned was defined as the angle between the original direction of the axon (initial angle) and a straight line connecting the base of the GC from the first to the last frame of the time-lapse recording. Axons with an initial angle <20° respect to the axis of the gradient were excluded, because they were already following the attractive gradient. Measurements were presented in rose histograms with bins of 15° and the length of each segment representing the frequency (in percent), average turned angle, and average displacement. Axon angles and outgrowth were measured with Fiji.
In utero electroporation
Pregnant CD1 females were anesthetized with intraperitoneal injections of a ketamine/xylazine mixture (75 mg/kg + 5 mg/kg body weight), their uterine horns were exposed, and the lateral ventricles of embryos were injected through the uterus wall using pulled capillaries (P-97; Sutter Instrument) filled with DNA preparations (2 µg/µl) diluted in PBS and colored with trypan blue. DNA-injected embryos were electroporated by using NEPA21 (30 v, pOn 50 ms, pOff 950 ms, five pulses; Nepagene) at E15-E15.5 (Jeanneteau et al., 2010). Postnatal mice were anesthetized on ice or with intraperitoneal injections of a ketamine/xylazine mixture and perfused intracardially with PBS followed by 4% PFA. Brains were removed, postfixed with 4% PFA for 1 wk, cryoprotected with 30% sucrose, and frozen in Tissue-Tek O.C.T (Sakura Finetek) at −80°C. Coronal brain cryosections of 60 µm thickness were obtained with a cryostate Microm HM550, and free-floating sections were processed for immunohistochemistry. The chicken anti-GFP antibody was used to label YFP-positive cells.
Immunofluorescence microscopy
Cultured neurons were fixed with 4% PFA/4% sucrose in PBS for 10 min at room temperature or with methanol/1 mM EGTA at −20°C for 5 min. For MT immunostaining (Witte et al., 2008), neurons were simultaneously permeabilized and fixed using 4% PFA/4% sucrose/0.25% glutaraldehyde/0.1% Triton X-100 diluted in PHEM buffer (60 mM Pipes, 25 mM Hepes, pH 7.4, 5 mM EGTA, and 1 mM MgCl2) for 10 min at room temperature. Fixed cells were permeabilized with 0.25% Triton X-100 in PBS for 5 min, blocked with 4% BSA (Sigma-Aldrich), and incubated in 2% BSA in PBS overnight with the primary antibodies detailed in the “Antibodies and drugs” section. F-actin was stained in fixed cells with phalloidin-TRITC (Sigma-Aldrich) according to the manufacturer’s instructions. For histological immunostainings, free-floating sections were incubated in 5% BSA/0.4% Triton X-100 in PBS for 2 h, followed by overnight primary antibody incubation in 2.5% BSA/0.4% Triton X-100 in PBS at 4°C. Alexa Fluor 350–, Alexa Fluor 488–, or Alexa Fluor 546–coupled secondary fluorescent antibodies (Life Technologies) were used at dilution 1:250 for 1 h. Nuclei were stained with DAPI, and samples were mounted using Prolong Gold Antifade (Molecular Probes). Cultured neurons and axonal GCs were imaged with the inverted confocal microscopes SP5-SMD (Leica) and LSM880 Airyscan (Zeiss), equipped with 63×/1.40 NA Oil objective. For MT imaging, confocal single-plane images of random axonal GCs from neuron cultures infected with lentiviruses were acquired using constant settings and avoiding signal saturation. A Zeiss Axioimager Z2 microscope equipped with 10×/0.3 NA Dry, 40×/1.4 NA Oil, and 100×/1.46 Oil objectives and a camera scMOS ZYLA 4.2 MP was used to image immunostained brain sections at the same rostrocaudal level of somatosensory cortex, HeLa cells, in vitro assays with fluorescent MTs and F-actin, and cortical cultures for mosaic generation of complete axons.
Time-lapse microscopy
Cortical cultures were plated in 35-mm diameter glass-bottom dishes (Ibidi); cotransfected at 1 DIV with the indicated reporters, reporters + shRNA constructs, or reporters + GFP-NAV1; and imaged 24 or 48 h later. In the rescue experiments, resistant GFP-NAV1 constructs were also cotransfected (2:1 shRNA/cDNA ratio). Live-imaging of MT dynamics, F-actin dynamics, and GC pathfinding was performed with an inverted microscope (Eclipse Ti; Nikon) equipped with a spinning disc (CSU-X1; Yokogawa) and an electron-multiplying charge-coupled device camera (iXon897; Andor). A 100×/1.45 NA Oil objective was used, adding an intermediate 1.5x lens in the SiR-tubulin imaging. In the GC motility experiments, a 40×/1.3 NA Oil objective was used. Isolated axonal GCs of random transfected neurons were imaged, except in NAV1 overexpression experiments where GFP-NAV1 comets and granules were visualized and therefore only splayed GCs were imaged. For this reason, GCs are overall larger and more dynamic in these overexpression experiments. In the rescue experiments, only GCs showing triple fluorescence signal were acquired. Image stacks were acquired using iQ3 software with a 0.5-µm step size at 1-s intervals for 2 min, at 1-s intervals for 40 s for SiR-Tubulin imaging, and at 1-min intervals for 20 min in the GC pathfinding experiments. For the analysis of parameters of MT dynamics in LLCPK-1α cells, cells were plated in glass-bottom 35-mm diameter dishes and transfected with the indicated mCherry-NAV1 constructs. After 24 h, live-cell imaging of mCherry-positive cells was performed on an inverted microscope (Eclipse Ti; Nikon) using a 100×/1.49 NA TIRF Oil objective, with an intermediate 1.5× lens, and an electron-multiplying charge-coupled device camera (Evolve 512; Photometrics). GFP-tagged α-tubulin was excited with a 488-nm argon laser line coupled to a motorized targeted laser illuminator (iLAS2; Roper Scientific). Excitation of mCherry was achieved with a Spectra-X LED Light Engine (Lumencor). Images were acquired at 1-s intervals for 2 min using Metamorph (Molecular Devices). All the cellular cultures were imaged at 37°C under a 5% CO2 atmosphere.
Image analysis
All images were processed and analyzed using Fiji software (National Institutes of Health). Before the analysis of videomicroscopy images, equal number of confocal planes were projected. The percentage of EB3-mCherry comets colocalizing with GFP-NAV1 and the GFP fluorescence mean gray values were measured in the first frame of the time lapses within the Lifeact-delineated areas of GCs and 10 µm upstream along the imaged axonal shaft. For all fluorescent intensity measurements, the background signal was measured in an adjacent area and subtracted. Analysis of the colocalization of endogenous NAV1, EB1, and MTs was performed on single-plane confocal images inside GCs and within 10 µm of axon shaft upstream the GC. Pearson’s coefficient was calculated using Coloc2 (Fiji) in the last frame of the time lapse before adding cytochalasin D and the first postdrug frame. Briefly, 8-bit images were background subtracted, and Pearson’s coefficient was calculated above thresholds to avoid zero-zero pixels. Analysis of NAV1 granules dynamics was performed in the GC periphery, including EB3-mCherry/GFP-NAV1 comets in which the EB3 fluorescence faded up. Retrograde translocating NAV1-granules were those that showed at least one phase of retrograde translocation. Paused NAV1-granules were those that remained static or showed retrograde translocation slower than 1.5 µm/min during the time-lapse. MT plus-end dynamics and F-actin RF analyses were performed using the kymograph plugin with Fiji (A. Seitz, European Molecular Biology Laboratory, Heidelberg, Germany), with lines drawn on the trajectory of GFP-NAV1 comets and granules or on the trajectory of fluorescently labeled MTs. For the analysis of NAV1 granules and RF speed in GCs, kymographs of both GFP-NAV1 and mCherry-actin signals were obtained from kymograph lines drawn on NAV1 granule trajectories. Actin RF speed values were obtained by averaging a minimum of three RF lines per kymograph. In the blebbistatin treatment experiments, mCherry-NAV1 particles and mCherry-Actin RF were imaged independently to avoid blebbistatin-induced phototoxicity due to 488-nm laser stimulation (Kolega, 2004). Kymographs in which NAV1 granules did not overlap with F-actin RF lines or run in the close vicinity were not included in the analysis. These cases represented 15% of the obtained kymographs. The parameters of MT dynamic instability in LLCPK-1α cells were determined by analyzing kymographs generated from TIRF time-lapse images. The length and duration of individual growth, catastrophe, and pausing events were obtained from manual fits. For calculation of instantaneous growth and shortening rates, the velocity of MT end displacements >0.5 µm were taken into account. The frequencies of transitions between phases were obtained by dividing the total number of transition events by the total time MTs spent either growing, shrinking, or pausing. For the MT density analysis in GCs, the mean fluorescence intensity of α-tubulin was measured in the phalloidin-positive areas of the GCs and ∼10 µm upstream along the imaged axonal trace. EB3-GFP and EB3-mCherry comet densities in GCs were obtained using an ImageJ custom-written macro (Sánchez-Huertas et al., 2016) and normalized to the F-actin–positive area outlined by the Lifeact fluorescence signal. EB3-mCherry and EB3-GFP comet polymerization speed, lifetime and length were measured inside Lifeact areas using plusTipTracker software (Applegate et al., 2011). The following tracking settings were used: search radius range, 5–12 pixels; minimum subtrack length, 3 frames; max gap length, 8 frames; max shrinkage factor, 0.8; max angle forward, 50; max angle backward, 10; fluctuation radius, 2.5 (Stout et al., 2014). EB3 comet analysis was limited to the first 60 s of the videos due to photobleaching thereafter. The quantification of the MT–F-actin colocalization in the in vitro crosslink assay was performed using an ImageJ custom-written macro kindly provided by E. Denarier (Elie et al., 2015). GC morphology was measured using Lifeact fluorescence in the first frame of the time-lapses. Filopodia were considered as thin protrusions with a uniform diameter ranging from 1–15 µm in length. GC perimeter was traced through the base of filopodia. For GC dynamics measurements, the filopodial extension events in each GC were tracked during the entire 2-min time lapses. In the GC motility experiments, GCs were classified as progressing when showing a net advance of at least 3 µm, as branching when the branches persisted during the total time of imaging, and bending when lines drawn on the axon shaft at frames 5 and 20 of the video formed an angle >20°. Only dynamic GCs extending at least one filopodium were included in the quantifications. For neuron radial migration analysis, YFP-expressing neurons were quantified in individual cortical layers or groups of cortical layers identified using nuclear staining, and the percentage of cells in each layer or group of layers was calculated. Layers I and II/III/IV were grouped as supragranular and layers V and VI as infragranular layers. Cell counts from three to four sections per animal were combined.
Statistical analysis
Statistical analysis was done using Prism 6 (GraphPad Software). The details are indicated in the figures and figure legends. Data distribution was assumed to be normal, but this was not formally tested. For all analyses, P values are represented as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Online supplemental material
Fig. S1 shows the localization of endogenous NAV1 at F-actin–rich areas and its colocalization with EB1 at MT plus ends in axonal GCs. Fig. S2 establishes that overexpression of NAV1 in cultured cortical neurons reduces GC size and dynamics. Fig. S3 reveals that NAV1 depletion impairs netrin-1–evoked axon branching. Fig. S4 depicts the experiments used to delineate the actin-binding domain of NAV1 with cytochalasin D–treated HeLa cells, and provides determination of its binding affinity (Kd) using an in vitro cosedimentation assay. Fig. S5 shows representative kymographs illustrating MT dynamics in LLCPK-1α overexpressing the WT and EBmut forms of mCherry-NAV1, as well as kymographs showing persistence of mCherry-NAV1 at the plus end of pausing MTs. Videos 1 and 2 reveal the morphology, dynamics and navigational behavior of the GCs of control and NAV1-depleted neurons. Video 3 illustrates netrin-1–evoked axon turning in the Dunn chamber assay. Video 4 shows the recruitment of GFP-NAV1 at MT plus ends in the F-actin–rich areas of GCs. Video 5 illustrates that GFP-NAV1-EBmut is impaired in MT plus-end tracking compared with GFP-NAV1-WT in GCs. Video 6 shows that mCherry-NAV1-WT, but not mCherry-NAV1-EBmut, tracks MT plus ends in LLCPK-1α cells. Video 7 shows an EB3-mCherry/GFP-NAV1 comet giving rise to a pausing EB3-negative NAV1 granule inside a GC filopodium. Video 8 shows the persistence of a GFP-NAV1 granule at the plus end of a SiR-tubulin–labeled nonpolymerizing MT within the F-actin–rich area of a GC.
Acknowledgments
We thank the staff of the Montpellier Ressources Imagerie facility for their assistance in microscopy, image analysis, statistical analyses, and rose histograms. We also thank the Plateforme de Vectorologie of Montpellier for the production and titering of lentiviral particles. We are grateful to Eric Denarier and Isabelle Arnal (Grenoble Institute of Neurosciences, Grenoble, France) for their invaluable help in the quantification of the F-actin–MT crosslinking assays. We are grateful to Irina Dudanova (Max Plank Institute of Neurobiology, Munich, Germany) for advice with the Dunn chamber assembly. We thank P. Wadsworth (University of Massachusetts, Boston, MA), B. Delaval (CRBM, Montpellier, France), J. Goedhart (University of Amsterdam, Netherlands), J. Lüders (IRB Barcelona, Barcelona, Spain), S. Bodin (CRBM, Montpellier, France), and D. Portran (CRBM, Montpellier, France) for providing reagents, and E. Sanchez for technical assistance. We also thank the members of the team for stimulating discussions and critical reading of the manuscript.
This work was supported by Agence Nationale de la Recherche (grant ANR-14-CE11-0025-01), Fondation pour la Recherche Médicale “Equipes FRM” program (grant DEQ20160334942), Fondation pour la Recherche Médicale “Projets innovants: financement d’un ingénieur” program (ING20150532167; A. Debant), and Centre National de la Recherche Scientifique (A. Debant and J. Boudeau). C. Fagotto-Kaufmann was funded by the University of Montpellier. F. Jeanneteau was funded by an Institut National de la Santé et de la Recherche Médicale AVENIR grant.
The authors declare no competing financial interests.
Author contributions: C. Sánchez-Huertas, J. Boudeau, and A. Debant designed the study and wrote the manuscript. C. Sánchez-Huertas and J. Boudeau conceived and executed the experimental work, and analyzed data. C. Sánchez-Huertas assembled the figures. M. Bonhomme performed experiments and analyzed data. A. Falco and C. Fagotto-Kaufmann assisted in various experiments and analyzed data. J. van Haren performed initial experiments on in vitro F-actin binding and analyzed data. F. Jeanneteau conceived and performed in utero electroporations and provided feedback. N. Galjart provided reagents and feedback and edited the manuscript. A. Debant secured funding and provided resources.
References
- Abe T., Yamazaki D., Murakami S., Hiroi M., Nitta Y., Maeyama Y., and Tabata T.. 2014. The NAV2 homolog Sickie regulates F-actin-mediated axonal growth in Drosophila mushroom body neurons via the non-canonical Rac-Cofilin pathway. Development. 141:4716–4728. 10.1242/dev.113308 [DOI] [PubMed] [Google Scholar]
- Akhmanova A., and Steinmetz M.O.. 2015. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16:711–726. 10.1038/nrm4084 [DOI] [PubMed] [Google Scholar]
- Alves-Silva J., Sánchez-Soriano N., Beaven R., Klein M., Parkin J., Millard T.H., Bellen H.J., Venken K.J., Ballestrem C., Kammerer R.A., et al. 2012. Spectraplakins promote microtubule-mediated axonal growth by functioning as structural microtubule-associated proteins and EB1-dependent +TIPs (tip interacting proteins). J. Neurosci. 32:9143–9158. 10.1523/JNEUROSCI.0416-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine-Bertrand J., Fu M., and Lamarche-Vane N.. 2016. Direct measurement of oscillatory RhoA activity in embryonic cortical neurons stimulated with the axon guidance cue netrin-1 using fluorescence resonance energy transfer. Biol. Cell. 108:115–126. 10.1111/boc.201500077 [DOI] [PubMed] [Google Scholar]
- Applegate K., Besson S., Matov A., Bagonis M., Jaqaman K., and Danuser G.. 2011. plusTipTracker: Quantitative Image Analysis Software for the Measurement of Microtubule Dynamics. J Struct Biol. 10.1016/j.jsb.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applewhite D.A., Grode K.D., Duncan M.C., and Rogers S.L.. 2013. The actin-microtubule cross-linking activity of Drosophila Short stop is regulated by intramolecular inhibition. Mol. Biol. Cell. 24:2885–2893. 10.1091/mbc.e12-11-0798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard G.J., Stear J.H., Noetzel T.L., Al-Bassam J., Kinoshita K., Harrison S.C., Howard J., and Hyman A.A.. 2008. XMAP215 is a processive microtubule polymerase. Cell. 132:79–88. 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarata G.M., Bearce E.A., and Lowery L.A.. 2016. Cytoskeletal social networking in the growth cone: How +TIPs mediate microtubule-actin cross-linking to drive axon outgrowth and guidance. Cytoskeleton (Hoboken). 73:461–476. 10.1002/cm.21272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis R. C., Ravindra Kumar S., Chan K. Y., Challis C., Beadle K., Jang M. J., Min Kim H., Rajendran P. S., Tompkins J. D., Shivkumar K., et al. 2019. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nature Protocols. 14:379–414. 10.1038/s41596-018-0097-3 [DOI] [PubMed] [Google Scholar]
- Coles C. H., and Bradke F.. 2015. Coordinating Neuronal Actin–Microtubule Dynamics. Current Biology. 25:R677–R691. 10.1016/j.cub.2015.06.020 [DOI] [PubMed] [Google Scholar]
- Coy J.F., Wiemann S., Bechmann I., Bächner D., Nitsch R., Kretz O., Christiansen H., and Poustka A.. 2002. Pore membrane and/or filament interacting like protein 1 (POMFIL1) is predominantly expressed in the nervous system and encodes different protein isoforms. Gene. 290:73–94. 10.1016/S0378-1119(02)00567-X [DOI] [PubMed] [Google Scholar]
- DeGeer J., Kaplan A., Mattar P., Morabito M., Stochaj U., Kennedy T.E., Debant A., Cayouette M., Fournier A.E., and Lamarche-Vane N.. 2015. Hsc70 chaperone activity underlies Trio GEF function in axon growth and guidance induced by netrin-1. J. Cell Biol. 210:817–832. 10.1083/jcb.201505084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent E. W., Barnes A. M., Tang F., and Kalil K.. 2004. Netrin-1 and semaphorin 3A promote or inhibit cortical axon branching, respectively, by reorganization of the cytoskeleton. J. Neurosci. 24:3002–3012. 10.1523/JNEUROSCI.4963-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent E.W., Gupton S.L., and Gertler F.B.. 2011. The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb. Perspect. Biol. 3 a001800 10.1101/cshperspect.a001800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon G.M., Tyler W.A., Omuro K.C., Kambouris J., Tyminski C., Henry S., Haydar T.F., Beffert U., and Ho A.. 2017. CLASP2 Links Reelin to the Cytoskeleton during Neocortical Development. Neuron. 93:1344–1358 e5. [DOI] [PMC free article] [PubMed]
- Dudanova I., Gatto G., and Klein R.. 2010. GDNF acts as a chemoattractant to support ephrinA-induced repulsion of limb motor axons. Curr. Biol. 20:2150–2156. 10.1016/j.cub.2010.11.021 [DOI] [PubMed] [Google Scholar]
- Elie A., Prezel E., Guérin C., Denarier E., Ramirez-Rios S., Serre L., Andrieux A., Fourest-Lieuvin A., Blanchoin L., and Arnal I.. 2015. Tau co-organizes dynamic microtubule and actin networks. Sci. Rep. 5:9964 10.1038/srep09964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassier C., Fréal A., Gasmi L., Delphin C., Ten Martin D., De Gois S., Tambalo M., Bosc C., Mailly P., Revenu C., et al. 2018. Motor axon navigation relies on Fidgetin-like 1-driven microtubule plus end dynamics. J. Cell Biol. 217:1719–1738. 10.1083/jcb.201604108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forscher P., and Smith S.J.. 1988. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J. Cell Biol. 107:1505–1516. 10.1083/jcb.107.4.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S., and Gordon-Weeks P.R.. 2009. Cytoskeletal dynamics in growth-cone steering. J. Cell Sci. 122:3595–3604. 10.1242/jcs.042309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S., Khanzada U.K., Parsons M., Chilton J.K., and Gordon-Weeks P.R.. 2008. Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat. Cell Biol. 10:1181–1189. 10.1038/ncb1778 [DOI] [PubMed] [Google Scholar]
- Gimona M., Djinovic-Carugo K., Kranewitter W.J., and Winder S.J.. 2002. Functional plasticity of CH domains. FEBS Lett. 513:98–106. 10.1016/S0014-5793(01)03240-9 [DOI] [PubMed] [Google Scholar]
- Hekimi S., and Kershaw D.. 1993. Axonal guidance defects in a Caenorhabditis elegans mutant reveal cell-extrinsic determinants of neuronal morphology. J. Neurosci. 13:4254–4271. 10.1523/JNEUROSCI.13-10-04254.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty-Ridilla J.L., Rankova A., Eskin J.A., Kenny K., and Goode B.L.. 2016. Accelerated actin filament polymerization from microtubule plus ends. Science. 352:1004–1009. 10.1126/science.aaf1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur E.M., Saijilafu B.D., Lee B.D., Kim S.J., Xu W.L., and Zhou F.Q.. 2011. GSK3 controls axon growth via CLASP-mediated regulation of growth cone microtubules. Genes Dev. 25:1968–1981. 10.1101/gad.17015911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins B.I., Li L., and Kalil K.. 2011. Wnt/calcium signaling mediates axon growth and guidance in the developing corpus callosum. Dev. Neurobiol. 71:269–283. 10.1002/dneu.20846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanneteau F., Deinhardt K., Miyoshi G., Bennett A.M., and Chao M.V.. 2010. The MAP kinase phosphatase MKP-1 regulates BDNF-induced axon branching. Nat. Neurosci. 13:1373–1379. 10.1038/nn.2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ka M., Jung E.M., Mueller U., and Kim W.Y.. 2014. MACF1 regulates the migration of pyramidal neurons via microtubule dynamics and GSK-3 signaling. Dev. Biol. 395:4–18. 10.1016/j.ydbio.2014.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn O.I., and Baas P.W.. 2016. Microtubules and Growth Cones: Motors Drive the Turn. Trends Neurosci. 39:433–440. 10.1016/j.tins.2016.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester M.P., Müller O., and Pollerberg G.E.. 2007. Adenomatous polyposis coli is differentially distributed in growth cones and modulates their steering. J. Neurosci. 27:12590–12600. 10.1523/JNEUROSCI.2250-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J. 2004. Phototoxicity and photoinactivation of blebbistatin in UV and visible light. Biochem. Biophys. Res. Commun. 320:1020–1025. 10.1016/j.bbrc.2004.06.045 [DOI] [PubMed] [Google Scholar]
- Kolodkin A.L., and Tessier-Lavigne M.. 2011. Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb. Perspect. Biol. 3 a001727 10.1101/cshperspect.a001727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Engel U., Rusch J., Scherrer S., Sheard K., and Van Vactor D.. 2004. The microtubule plus end tracking protein Orbit/MAST/CLASP acts downstream of the tyrosine kinase Abl in mediating axon guidance. Neuron. 42:913–926. 10.1016/j.neuron.2004.05.020 [DOI] [PubMed] [Google Scholar]
- Leung C.L., Sun D., Zheng M., Knowles D.R., and Liem R.K.. 1999. Microtubule actin cross-linking factor (MACF): a hybrid of dystonin and dystrophin that can interact with the actin and microtubule cytoskeletons. J. Cell Biol. 147:1275–1286. 10.1083/jcb.147.6.1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., and Dwyer T.. 2014. Microtubule dynamics in axon guidance. Neurosci. Bull. 30:569–583. 10.1007/s12264-014-1444-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie R.K., Bahmanyar S., Siemers K.A., Votin V., Chang P., Stearns T., Nelson W.J., and Barth A.I.M.. 2004. Adenomatous polyposis coli and EB1 localize in close proximity of the mother centriole and EB1 is a functional component of centrosomes. J. Cell Sci. 117:1117–1128. 10.1242/jcs.00939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes T., Barceló A., and Buesa C.. 2002. Neuron navigator: a human gene family with homology to unc-53, a cell guidance gene from Caenorhabditis elegans. Genomics. 80:21–30. 10.1006/geno.2002.6799 [DOI] [PubMed] [Google Scholar]
- Marcus-Gueret N., Schmidt K.L., and Stringham E.G.. 2012. Distinct cell guidance pathways controlled by the Rac and Rho GEF domains of UNC-73/TRIO in Caenorhabditis elegans. Genetics. 190:129–142. 10.1534/genetics.111.134429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-López M.J., Alcántara S., Mascaró C., Pérez-Brangulí F., Ruiz-Lozano P., Maes T., Soriano E., and Buesa C.. 2005. Mouse neuron navigator 1, a novel microtubule-associated protein involved in neuronal migration. Mol. Cell. Neurosci. 28:599–612. 10.1016/j.mcn.2004.09.016 [DOI] [PubMed] [Google Scholar]
- McNeill E.M., Roos K.P., Moechars D., and Clagett-Dame M.. 2010. Nav2 is necessary for cranial nerve development and blood pressure regulation. Neural Dev. 5:6 10.1186/1749-8104-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill E.M., Klöckner-Bormann M., Roesler E.C., Talton L.E., Moechars D., and Clagett-Dame M.. 2011. Nav2 hypomorphic mutant mice are ataxic and exhibit abnormalities in cerebellar development. Dev. Biol. 353:331–343. 10.1016/j.ydbio.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill R.A., Plum L.A., Kaiser M.E., and Clagett-Dame M.. 2002. A mammalian homolog of unc-53 is regulated by all-trans retinoic acid in neuroblastoma cells and embryos. Proc. Natl. Acad. Sci. USA. 99:3422–3427. 10.1073/pnas.052017399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen K., and Larsson L.I.. 2003. Effects of cytochalasin D on the actin cytoskeleton: association of neoformed actin aggregates with proteins involved in signaling and endocytosis. Cell. Mol. Life Sci. 60:1007–1012. 10.1007/s00018-003-3022-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muley P.D., McNeill E.M., Marzinke M.A., Knobel K.M., Barr M.M., and Clagett-Dame M.. 2008. The atRA-responsive gene neuron navigator 2 functions in neurite outgrowth and axonal elongation. Dev. Neurobiol. 68:1441–1453. 10.1002/dneu.20670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukirchen D., and Bradke F.. 2011. Cytoplasmic linker proteins regulate neuronal polarization through microtubule and growth cone dynamics. J. Neurosci. 31:1528–1538. 10.1523/JNEUROSCI.3983-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Bartolini F., Deaconescu A.M., Moseley J.B., Dogic Z., Grigorieff N., Gundersen G.G., and Goode B.L.. 2010. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J. Cell Biol. 189:1087–1096. 10.1083/jcb.201001016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Yadav V., Sharma A., Khurana J.P., and Pandey G.K.. 2018. The unc-53 gene negatively regulates rac GTPases to inhibit unc-5 activity during Distal tip cell migrations in C. elegans. Cell Adhes. Migr. 12:195–203. 10.1080/19336918.2017.1345413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters P.J., Baker A., Goris I., Daneels G., Verhasselt P., Luyten W.H., Geysen J.J., Kass S.U., and Moechars D.W.. 2004. Sensory deficits in mice hypomorphic for a mammalian homologue of unc-53. Brain Res. Dev. Brain Res. 150:89–101. 10.1016/j.devbrainres.2004.03.004 [DOI] [PubMed] [Google Scholar]
- Purro S.A., Ciani L., Hoyos-Flight M., Stamatakou E., Siomou E., and Salinas P.C.. 2008. Wnt regulates axon behavior through changes in microtubule growth directionality: a new role for adenomatous polyposis coli. J. Neurosci. 28:8644–8654. 10.1523/JNEUROSCI.2320-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinson D. A., Dillon C. P., Kwiatkowski A. V., Sievers C., Yang L., Kopinia J., Rooney D. L., Zhang M., Ihrig M. M., McManus M. T., et al. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nature Genetics. 33:401–406. [DOI] [PubMed] [Google Scholar]
- Rusan N.M., Fagerstrom C.J., Yvon A.M., and Wadsworth P.. 2001. Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol. Biol. Cell. 12:971–980. 10.1091/mbc.12.4.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Huertas C., Freixo F., Viais R., Lacasa C., Soriano E., and Lüders J.. 2016. Non-centrosomal Nucleation Mediated by Augmin Organizes Microtubules in Post-Mitotic Neurons and Controls Axonal Microtubule Polarity. Nat Comm. 7:12187 10.1038/ncomms12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A.W., Schoonderwoert V.T.G., Ji L., Mederios N., Danuser G., and Forscher P.. 2008. Coordination of actin filament and microtubule dynamics during neurite outgrowth. Dev. Cell. 15:146–162. 10.1016/j.devcel.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K.L., Marcus-Gueret N., Adeleye A., Webber J., Baillie D., and Stringham E.G.. 2009. The cell migration molecule UNC-53/NAV2 is linked to the ARP2/3 complex by ABI-1. Development. 136:563–574. 10.1242/dev.016816 [DOI] [PubMed] [Google Scholar]
- Slater P.G., Cammarata G.M., Samuelson A.G., Magee A., Hu Y., and Lowery L.A.. 2019. XMAP215 promotes microtubule-F-actin interactions to regulate growth cone microtubules during axon guidance in Xenopuslaevis. J. Cell Sci. 132 jcs224311 10.1242/jcs.224311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T., Slemmer J., Hoogenraad C.C., Lansbergen G., Dortland B., De Zeeuw C.I., Grosveld F., van Cappellen G., Akhmanova A., and Galjart N.. 2003. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 23:2655–2664. 10.1523/JNEUROSCI.23-07-02655.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout A., D’Amico S., Enzenbacher T., Ebbert P., and Lowery L.A.. 2014. Using plusTipTracker Software to Measure Microtubule Dynamics in Xenopus Laevis Growth Cones. J Vis Exp. 91:e52138 10.3791/52138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradal T., Kranewitter W., Winder S.J., and Gimona M.. 1998. CH domains revisited. FEBS Lett. 431:134–137. 10.1016/S0014-5793(98)00751-0 [DOI] [PubMed] [Google Scholar]
- Stringham E.G., and Schmidt K.L.. 2009. Navigating the cell: UNC-53 and the navigators, a family of cytoskeletal regulators with multiple roles in cell migration, outgrowth and trafficking. Cell Adhes. Migr. 3:342–346. 10.4161/cam.3.4.9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringham E., Pujol N., Vandekerckhove J., and Bogaert T.. 2002. unc-53 controls longitudinal migration in C. elegans. Development. 129:3367–3379. [DOI] [PubMed] [Google Scholar]
- Tsvetkov A.S., Samsonov A., Akhmanova A., Galjart N., and Popov S.V.. 2007. Microtubule-binding proteins CLASP1 and CLASP2 interact with actin filaments. Cell Motil. Cytoskeleton. 64:519–530. 10.1002/cm.20201 [DOI] [PubMed] [Google Scholar]
- van Haren J., Draegestein K., Keijzer N., Abrahams J.P., Grosveld F., Peeters P.J., Moechars D., and Galjart N.. 2009. Mammalian Navigators are microtubule plus-end tracking proteins that can reorganize the cytoskeleton to induce neurite-like extensions. Cell Motil. Cytoskeleton. 66:824–838. 10.1002/cm.20370 [DOI] [PubMed] [Google Scholar]
- van Haren J., Boudeau J., Schmidt S., Basu S., Liu Z., Lammers D., Demmers J., Benhari J., Grosveld F., Debant A., et al. 2014. Dynamic microtubules catalyze formation of navigator-TRIO complexes to regulate neurite extension. Curr. Biol. 24:1778–1785. 10.1016/j.cub.2014.06.037 [DOI] [PubMed] [Google Scholar]
- Wang P., Yang X., Wu P., Zhang J., Sato T., Yamagata S., and Yamagata T.. 2007. GM3 signals regulating TNF-alpha expression are mediated by Rictor and Arhgdib in mouse melanoma B16 cells. Oncology. 73:430–438. [DOI] [PubMed] [Google Scholar]
- Watson P., and Stephens D. J.. 2006. Microtubule plus-end loading of p150Glued is mediated by EB1 and CLIP-170 but is not required for intracellular membrane traffic in mammalian cells. Journal of Cell Science. 119:2758–2767. 10.1242/jcs.02999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S.R., and Lauring B.. 2007. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 8:1657–1667. 10.1111/j.1600-0854.2007.00642.x [DOI] [PubMed] [Google Scholar]
- Witte H., Neukirchen D., and Bradke F.. 2008. Microtubule Stabilization Specifies Initial Neuronal Polarization. Journal of Cell Biology. 180(3):619–632. 10.1083/jcb.200707042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam P.T., Langlois S.D., Morin S., and Charron F.. 2009. Sonic hedgehog guides axons through a noncanonical, Src-family-kinase-dependent signaling pathway. Neuron. 62:349–362. 10.1016/j.neuron.2009.03.022 [DOI] [PubMed] [Google Scholar]
- Zhang J.-H., Zhao Y.-F., He X.-X., Zhao Y., He Z.-X., Zhang L., Huang Y., Wang Y.-B., Hu L., Liu L., et al. 2018. DCC-Mediated Dab1 Phosphorylation Participates in the Multipolar-to-Bipolar Transition of Migrating Neurons. Cell Rep. 22:3598–3611. 10.1016/j.celrep.2018.03.005 [DOI] [PubMed] [Google Scholar]
- Zhou F.Q., Zhou J., Dedhar S., Wu Y.H., and Snider W.D.. 2004. NGF-induced axon growth is mediated by localized inactivation of GSK-3beta and functions of the microtubule plus end binding protein APC. Neuron. 42:897–912. 10.1016/j.neuron.2004.05.011 [DOI] [PubMed] [Google Scholar]