Abstract
Arginines within the third intracellular loop of the C. elegans OCTR-1 and human ADRA2A receptors are methylated by the human protein arginine methyltransferase PRMT5 in vitro . Methylation of these residues could serve to modulate receptor signaling in vivo .
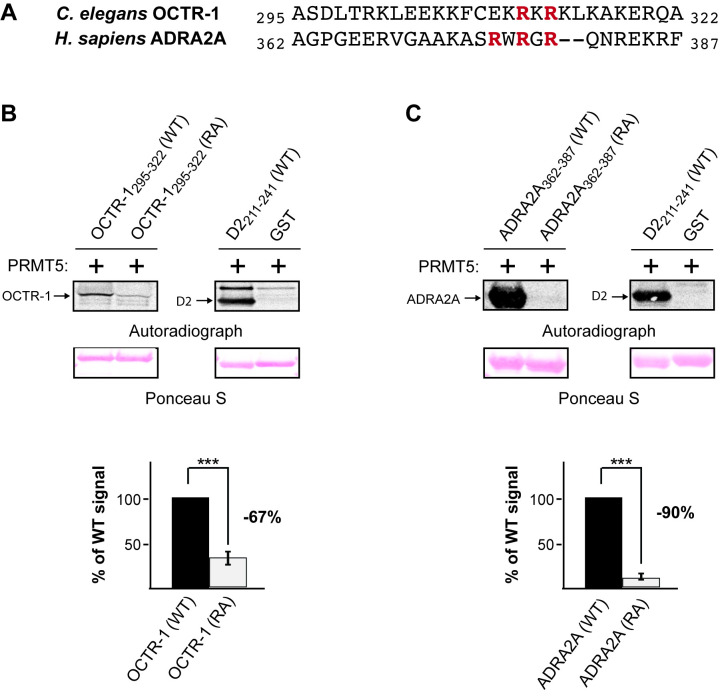
Figure 1. Human PRMT5 methylates the third intracellular loop of the C. elegans OCTR-1 receptor and the human alpha-2A adrenergic receptor (ADRA2A) in vitro .
(A) Alignment showing conservation of the predicted arginine methylation motifs in the C. elegans OCTR-1 and human ADRA2A receptors. The entire third intracellular loop (3 rd ICL) of OCTR-1 is predicted to have ~121 amino acids, representing residues 203-323 [SWISS-MODEL, based on (Qu et al. 2020)]; only residues 295-322 of the 3 rd ICL are shown and aligned with the corresponding residues of the 3 rd ICL of ADRA2A. Arginines within the predicted methylation motifs (RXR) are shown in red. (B and C) Representative blots for the in vitro methylation assays. A wild-type (WT) and mutant (RA) recombinant fragment of the 3 rd ICL of OCTR-1 (residues 295-322) and ADRA2A (residues 362-387), flanked both amino- and carboxy-terminally with an S-tag to increase solubility, and fused to glutathione S-transferase (GST) were used in an in vitro methylation assay with active recombinant human PRMT5. There are no arginines within the S-tag. A fragment of the human D2 dopamine receptor was used as the positive control for methylation (Likhite et al. 2015) and GST alone served as the negative control. Top: Autoradiographs show that WT OCTR-1 and ADRA2A are methylated by PRMT5. Fragments with the conserved arginines (shown in red, Figure 1A) changed to alanines were not efficiently methylated. Ponceau S staining of the polyvinylidene difluoride (PVDF) membranes was performed to demonstrate equivalent loading of the WT and RA receptor fragments. Bottom: Quantification of the degree of methylation of the receptor fragments based on densitometric analysis of the autoradiographs. Loss of the methylation motif resulted in a 67% decrease in OCTR-1 methylation and a 90% decrease in ADRA2A methylation. Data are means, +/- the standard error of the mean (SEM) from three independent experiments. *** = p < 0.001
Description
One way that G protein-coupled signal transduction is regulated intracellularly is through post-translational modification of the receptors (G protein-coupled receptors; GPCRs). While phosphorylation remains the best studied of these modifications (Komolov and Benovic 2018), a proteomics analysis previously identified several GPCRs as substrates of an anti-methylarginine antibody (Boisvert et al. 2003). We subsequently reported the first evidence that GPCRs can be functionally regulated by protein arginine methylation: D2-like dopamine receptors (human D2 and C. elegans DOP-3 receptors) (Likhite et al. 2015) and the C. elegans SER-2 tyramine receptor (Bowitch et al. 2018). In each case, conserved arginines within the third intracellular loop (3 rd ICL) were methylated by PRMT5 in vitro , and experiments examining signaling in cell culture (D2) and behavior in C. elegans (DOP-3 and SER-2) suggested that arginine methylation serves to promote receptor signaling (Likhite et al. 2015; Bowitch et al. 2018). However, the mechanism by which it does so is not yet known. In the case of D2, methylation may alter interaction with regulatory proteins that bind to the region of the 3 rd ICL that contains the methylated arginines (Bowitch et al. 2021).
In a bioinformatics analysis, Likhite et al. (2015) identified 300 human GPCRs that contained an intracellular putative methylation motif [RGG or RXR, where X can be any amino acid (Bedford and Clarke 2009; Thandapani et al. 2013)]. Nearly 70% of these motifs were conserved in the corresponding mouse and rat receptors, and ~1% (seven) were conserved in C. elegans (Likhite et al. 2015). Among these, the C. elegans OCTR-1 octopamine receptor was identified to contain a putative methylation motif that is conserved in the human alpha-2A adrenergic receptor (ADRA2A) (Likhite et al. 2015). Octopamine was first discovered in the salivary glands of the octopus in 1948 (Juorio and Molinoff 1974), and has since been determined to be a biogenic amine in invertebrates that functions as a neurotransmitter, neurohormone and neuromodulator (Scheiner et al. 2002). It is considered the invertebrate counterpart of adrenaline due to their structural and receptor similarity (Roeder 1999). Adrenaline and noradrenaline (with a lower potency) are endogenous ligands of ADRA2A (Altosaar et al. 2021).
We wished to determine if the predicted methylation motif within the 3 rd ICL of OCTR-1 and ADRA2A (Figure 1A) could serve as a substrate for human PRMT5 in vitro . A recombinant fragment of the 3 rd ICL of each (see Figure Legend for details) was methylated by PRMT5 (Figure 1B, 1C). To determine whether the arginines of the predicted methylation motif of OCTR-1 (Arg 311 and Arg 313 ) were necessary for methylation, we generated a recombinant fragment in which these arginines were changed to alanine [OCTR-1(RA)]. Methylation of this fragment was decreased 67% compared to the wild-type (WT) fragment (Figure 1B). Similarly, to determine whether the arginines of the predicted methylation motif of ADRA2A (Arg 376 , Arg 378 and Arg 380 ) were necessary for methylation, we generated a recombinant fragment in which these arginines were changed to alanine [ADRA2A(RA)]. Methylation of this fragment was decreased 90% compared to the WT fragment (Figure 1C). Both peptides contain additional arginines that are not part of described PRMT recognition motifs. Context-dependent methylation of these could account for the residual signals, as PRMT5 is not known to methylate lysines. These data establish the 3 rd ICL of both receptors as substrates for PRMT5 in vitro , and suggest that the highlighted arginines are key sites of methylation within this region.
GPCRs are the largest family of cell surface receptors and are involved in diverse physiological processes. Accordingly, mutations that disrupt GPCR signaling can result in many different diseases (Schöneberg et al. 2004; Zalewska et al. 2014). Nearly 35% of all pharmaceuticals target GPCRs in an attempt to therapeutically control their signaling (Sriram and Insel 2018). Notably, arginine methylation appears to have a modulatory effect on GPCR signaling (Likhite et al. 2015; Bowitch et al. 2018). Thus, identifying additional receptors that can be methylated provides new potential targets for treatments based on regulating or altering GPCR methylation status.
Methods
Protein purification
The following GPCR third intracellular loop fragments were purified using the previously described protocol (Bowitch et al. 2018):
GST::S-tag::OCTR-1 295-322 ::S-tag
GST::S-tag::OCTR-1 295-322 (R311A/R313A)::S-tag
GST::S-tag::ADRA2A 362-387 ::S-tag
GST::S-tag::ADRA2A 362-387 (R376A/R378A/R380A)::S-tag
The S-tag sequence (KETAAAKFERQHMDS) was added to aid in protein solubility and does not contain any arginines.
GST and GST::D2 211-241 were expressed and purified as previously described (Likhite et al. 2015).
In vitro methylation
The in vitro methylation assay was performed similarly to as described (Likhite et al. 2015; Bowitch et al. 2018) in a total volume of 20 µl with 4 µg of substrate, 1 µl of recombinant human PRMT5+MEP50 protein (Abcam; catalog number 271720), and 2 µCi of S-[methyl- 3 H]adenosyl-L-methionine (55 to 85 Ci/mmol; PerkinElmer) in 20 mM phosphate buffer (pH 7.4). Reactions were incubated at 30°C for 4 hours, resolved by Bolt 4-20% SDS-polyacrylamide gel electrophoresis (Thermo-Fisher), and then transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were stained with Ponceau S, dried, and exposed to a low energy phosphorimager screen for at least two weeks and subsequently developed using an Azure Sapphire Biomolecular Imager. Band intensities were quantified with Bio-Rad ImageLab software (6.1.0 build 7) and were normalized according to gel loading (Ponceau S stained bands quantified by ImageJ, version 1.53n). The Student’s one-tailed t-Test was used for statistical analysis.
Acknowledgments
Acknowledgments
this.convertHTML(ack)
Funding
This work was supported by the National Institutes of Health (R21MH101386 and R01DC015758 to DMF).
References
- Altosaar K, Balaji P, Bond RA, Bylund DB, Cotecchia S, Devost D, Doze VA, Eikenburg DC, Gora S, Goupil E., Graham RM, Hébert T, Hieble JP, Hills R, Kan S, Machkalyan G, Michel MC, Minneman KP, Parra S, Perez D, Sleno R, Summers R, Zylbergold P. 2021. Adrenoceptors in GtoPdb v.2021.3. IUPHAR/BPS Guide to Pharmacology CITE 2021.
- Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009 Jan 16;33(1):1–13}. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Côté J, Boulanger MC, Richard S. A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics. 2003 Oct 7;2(12):1319–1330}. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- Bowitch A, Michaels KL, Yu MC, Ferkey DM. The Protein Arginine Methyltransferase PRMT-5 Regulates SER-2 Tyramine Receptor-Mediated Behaviors in Caenorhabditis elegans . . G3 (Bethesda) 2018 Jul 2;8(7):2389–2398}. doi: 10.1534/g3.118.200360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowitch A, Sahoo A, Clark AM, Ntangka C, Raut KK, Gollnick P, Yu MC, Pascal SM, Walker SE, Ferkey DM. Methylation of the D2 dopamine receptor affects binding with the human regulatory proteins Par-4 and Calmodulin. MicroPubl Biol. 2021 Feb 9;2021 doi: 10.17912/micropub.biology.000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juorio AV, Molinoff PB. The normal occurrence of octopamine in neural tissue of the Octopus and other cephalopods. J Neurochem. 1974 Feb 1;22(2):271–280}. doi: 10.1111/j.1471-4159.1974.tb11590.x. [DOI] [PubMed] [Google Scholar]
- Komolov KE, Benovic JL. G protein-coupled receptor kinases: Past, present and future. Cell Signal. 2017 Jul 12;41:17–24}. doi: 10.1016/j.cellsig.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likhite N, Jackson CA, Liang MS, Krzyzanowski MC, Lei P, Wood JF, Birkaya B, Michaels KL, Andreadis ST, Clark SD, Yu MC, Ferkey DM. The protein arginine methyltransferase PRMT5 promotes D2-like dopamine receptor signaling. Sci Signal. 2015 Nov 10;8(402):ra115–ra115. doi: 10.1126/scisignal.aad0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L, Zhou QT, Wu D, Zhao SW. 2020. Crystal structures of the alpha2A adrenergic receptor in complex with an antagonist RSC. doi:10.2210/pdb6KUX/pdb.
- Roeder T. Octopamine in invertebrates. Prog Neurobiol. 1999 Dec 1;59(5):533–561}. doi: 10.1016/s0301-0082(99)00016-7. [DOI] [PubMed] [Google Scholar]
- Scheiner R, Plückhahn S, Oney B, Blenau W, Erber J. Behavioural pharmacology of octopamine, tyramine and dopamine in honey bees. Behav Brain Res. 2002 Nov 15;136(2):545–553}. doi: 10.1016/s0166-4328(02)00205-x. [DOI] [PubMed] [Google Scholar]
- Schöneberg T, Schulz A, Biebermann H, Hermsdorf T, Römpler H, Sangkuhl K. Mutant G-protein-coupled receptors as a cause of human diseases. Pharmacol Ther. 2004 Dec 1;104(3):173–206}. doi: 10.1016/j.pharmthera.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Sriram K, Insel PA. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol Pharmacol. 2018 Jan 3;93(4):251–258}. doi: 10.1124/mol.117.111062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thandapani P, O'Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol Cell. 2013 Jun 6;50(5):613–623}. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- Zalewska M, Siara M, Sajewicz W. G protein-coupled receptors: abnormalities in signal transmission, disease states and pharmacotherapy. Acta Pol Pharm. 2014 Apr 1;71(2):229–243}. [PubMed] [Google Scholar]