Abstract
Strains of enterohemorrhagic Escherichia coli (EHEC) serotype O157:H7 produce under stress copious amounts of exopolysaccharide (EPS) composed of colanic acid (CA). Studies were performed to evaluate the association of production of CA with survival of EHEC under adverse environmental conditions. A CA-deficient mutant, M4020, was obtained from a CA-proficient parental strain, E. coli O157:H7 W6-13, by inserting a kanamycin resistance gene cassette (kan) into wcaD and wcaE, 2 of the 21 genes required for CA biosynthesis. M4020 was defective in CA production as determined from the ratio of uronic acid to protein (UA/P) of cells grown from 1 to 4 days at 25°C on minimal glucose agar (MGA), MacConkey agar, and sorbitol-MacConkey agar, and by colony morphology on MGA. The results of stress treatment revealed that M4020 was substantially less tolerant to acid (pH 4.5 and 5.5) and heat (55 and 60°C) in comparison to W6-13, indicating that CA confers on E. coli O157:H7 a protective effect from the environmental stresses of acid and heat.
Escherichia coli O157:H7 is a serious foodborne pathogen, causing life-threatening maladies including hemorrhagic colitis, hemolytic-uremic syndrome, and thrombotic-thrombocytopenic purpura (22, 25, 30). Although outbreaks of E. coli O157:H7 infection are frequently associated with eating undercooked ground beef, a variety of other foods, including dry and acidic foods, also have been implicated as vehicles of infection (4, 5). Outbreaks associated with highly acidic foods are of particular concern because acidic conditions are normally considered sufficient not only to inhibit the growth of but also to kill most foodborne pathogens. Hence, the tolerance of E. coli O157:H7, which has a low infectious dose, to acidic foods compounds the serious nature of this bacterium as a foodborne pathogen.
Many strains of E. coli O157:H7 are substantially more tolerant to adverse environmental conditions than many nonpathogenic E. coli strains (2, 17), with the pathogen surviving for prolonged periods of time in a range of foods under various adverse conditions (20, 26). E. coli O157:H7 survived acid stress in 1.5% (vol/vol) acetic acid (pH 4.0) at 37°C for 15 min, whereas a nonpathogenic strain of E. coli did not survive under the same conditions (32). In a laboratory study, E. coli O157:H7 survived the drying process used to make venison jerky under several time and temperature combinations, including up to 10 h at 62.8°C (15). Viable E. coli O157:H7 was detected after 158 days of ripening in cheddar cheese made with milk inoculated with 103 CFU/ml (24). While the endurance of E. coli O157:H7 to environmental stress has been well documented, the molecular mechanisms affecting such tolerances are poorly understood.
Many factors contribute to bacterial resistance to environmental stress. Among these are cell surface structures and appendages which provide the firstline of defense for a bacterium. As with many other members of the Enterobacteriaceae, E. coli secretes a variety of exopolysaccharides (EPS), including colanic acid (CA). CA contains l-fucose, d-glucuronic acid, d-glucose, d-galactose, and pyruvate and forms a thick mucoid matrix on cell surfaces (8, 10, 23, 31). CA biosynthesis in E. coli K-12 is encoded by the wca gene cluster, which includes 21 open reading frames (ORFs) (28). Among these ORFs, wcaC and wcaE encode putative CA glycosyl transferases, and wcaD produces a putative CA polymerase. These three genes are located on the upper portion of the giant CA operon and are involved in sequential transfer of the component sugars of CA and assembly of the CA polymer.
Junkins and Doyle examined 27 E. coli O157:H7 strains and determined that 67% (18 of 27) formed mucoid colonies on sorbitol-MacConkey (SMAC) agar during prolonged incubation at ambient temperature (14). Four of the five nonslime-forming strains on SMAC agar did become mucoid on media containing a higher concentration of sodium chloride. Further analysis revealed that CA was the principal component of EPS produced by E. coli O157:H7. However, no association has been made with CA and its influence on survival of E. coli O157:H7 under conditions of stress.
In the study presented here, a CA-deficient mutant was constructed by inserting a kanamycin resistance gene cassette (kan) into the wca operon of the E. coli O157:H7 genome. CA production by the parental and mutant strains was determined by measurement of the uronic acid/protein (UA/P) ratio (14). Heat or acid treatments were subsequently applied to the parental and mutant strains to determine the survival characteristics of the isogenic pair.
Construction of CA-deficient mutant.
More than a hundred E. coli O157:H7 strains from our laboratory collection were screened on SMAC agar for EPS-forming colonies, a presumptive indication of CA production. CA-producing E. coli O157:H7 W6-13 was eventually selected as a parental strain because of its production of copious amounts of EPS. Two PCR fragments (upstream and downstream) were amplified from the W6-13 chromosome using the oligonucleotide primer pairs, P1 and P2 (5′-AAAGCTTAAACCGGACGTCACT-3′ and 5′-GAATTCTCCTCCACACCATGCCAAT-3′; EcoRI site underlined) and P3 and P4 (5′-AAAGAATTCCAAAGGCATTGC-3′ and 5′-TTCGCGTCAGCACACAATTC-3′; EcoRI site underlined), respectively. The PCR primers were derived based on GenBank sequences within wcaC, wcaD, and wcaE of E. coli K-12 (GenBank accession no. U38473). Both primers 2 and 3 had an artificial EcoRI site near the 5′ end. Following PCR, the two amplified DNA fragments were digested with EcoRI and then ligated to each other to create an EcoRI restriction site. This fragment with the EcoRI site was subsequently cloned into plasmid vector pGEM-T (Promega Co., Madison, Wis.) to generate recombinant plasmid pYM37 (wcaΔDΔE). The EcoRI restriction site on pYM37 was used to receive an EcoRI-EcoRI kanamycin resistance gene cassette which was PCR amplified from pNEO (Amersharm Pharmacia Biotech, Inc., Piscataway, N.J.) and restricted with EcoRI. The resulting plasmid was designated pYM3720 from which the wcaDE::kan fragment was amplified with primer pair P1 and P4. Amplified wcaDE::kan fragment was introduced into E. coli O157:H7 W6-13 by electroporation. Recombinant colonies were selected on Luria-Bertani agar supplemented with kanamycin (100 μg/ml). One of the colonies, M4020 (W6-13 wcaDE::kan), was isolated. The site of insertion was confirmed by PCR amplification and Southern hybridization with a kanamycin resistance gene probe. Insertion mutagenesis abolished the function of wca, whereby M4020 became defective in CA production.
wcaC, wcaD, and wcaE of E. coli O157:H7 were sequenced using an ABI Prism PCR Sequencer (Molecular Genetics Instrumentation Facility, University of Georgia). The sequences were deposited in GenBank (accession no. AF320069). Sequence analysis revealed that these genes share 98% homology with the same genes of E. coli K-12.
Quantification of CA production.
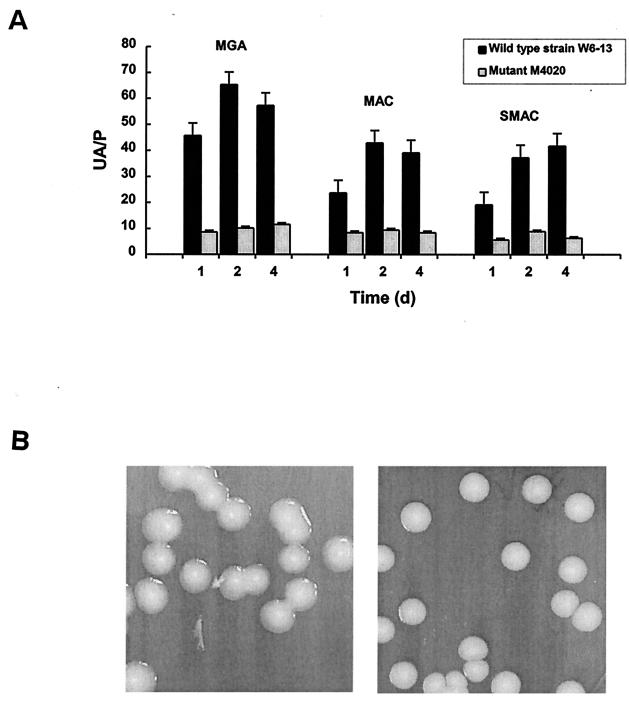
CA produced by E. coli W6-13 and M4020 was quantified according to a procedure previously described by Junkins and Doyle (14). The UA/P ratio, defined as micrograms of uronic acid per milligram of protein, was used to estimate CA production of both strains on minimal glucose agar (MGA), MacConkey (MAC) agar, and SMAC agar. While colonies of W6-13 and M4020 were visible on agar plates after 16 h of incubation, development of a mucoid appearance by W6-13 required at least 24 h of incubation at 25°C. UA/P ratios of the same strain varied substantially among media used for cultivating the cells. However, CA production increased with incubation time, with the greatest amount being produced at day 2 on MGA and MAC agar and at day 4 on SMAC among the various days of incubation studied (Fig. 1A). Colonies became extremely mucoid due to overproduction of CA (Fig. 1B). Greater amounts of CA were observed when W6-13 was grown on MGA than on complex media, such as MAC agar, SMAC agar (Fig. 1A), and MGA containing 0.5% Casamino Acids (wt/vol) (data not shown). This agrees with previous reports that CA production in E. coli K-12 is enhanced when cells are grown under less-than-optimal conditions for growth, such as in presence of low temperature, high salt concentrations, or high concentrations of utilizable carbon sources (18, 21, 27). Although many strains of E. coli O157:H7 produce CA, the amounts produced can vary greatly. Furthermore, CA production occurs when cultures grow at 25°C rather than at 37°C, which may be a feature to provide bacteria with a means to respond to stress and better survive conditions outside their human and animal hosts.
FIG. 1.
UA/P ratio of E. coli O157:H7 W6-13 and its CA-deficient mutant M4020 when grown on different media, and CA production by colonies of W6-13 and M4020. (A) UA/P ratio of W6-13 and M4020 grown at 25°C for 1, 2, and 4 days on MGA, MAC agar, and SMAC agar. (B) CA production of E. coli O157:H7 W6-13 (left) and M4020 (right) when grown on MGA at 25°C for 4 days.
CA production by E. coli O157:H7 W6-13 was also compared to that of the nonpathogenic E. coli K-12 strain ZK2686 which is known to produce CA (6). The K-12 strain produced colonies with much less EPS than by E. coli O157:H7 W6-13 under the same conditions on MGA, supporting our earlier hypothesis that the overproduction of CA may play a more important role in greater resistance to environmental stresses by certain pathogenic than nonpathogenic microorganisms.
Protective effect of CA on acid and thermal tolerance.
In this study, W6-13 and M4020 were subjected to acid treatment with pH levels ranging from 4.5 to 6.5 and to heat treatment at 55 and 60°C to evaluate the effect of CA expression on acid and thermal tolerance. For the acid treatment, W6-13 and M4020 were inoculated onto MGA and incubated at 25°C for 24 h. Colonies were removed with minimal glucose broth (MGB), and cell suspensions of W6-13 and M4020 were inoculated (1%) (vol/vol) into fresh MGB containing 5% Casamino Acids adjusted to pH 4.5, 5.5, and 6.5 with 0.1 M HCl. The cultures were incubated at 37°C, and viable counts of E. coli O157:H7 were determined at selected intervals. Approximately 108 CFU/ml of cell suspension of each strain was heated at 55 and 60°C for the thermal inactivation studies. Viable counts of E. coli O157:H7 were determined by sampling at selected time intervals, plating bacteria on MGA containing 0.5% Casamino Acids and incubating the plates at 37°C for 24 h. The D-value, which is defined as the time at a specific temperature that is required to inactivate 1 log of the bacterial population (12), was calculated by linear regression analysis.
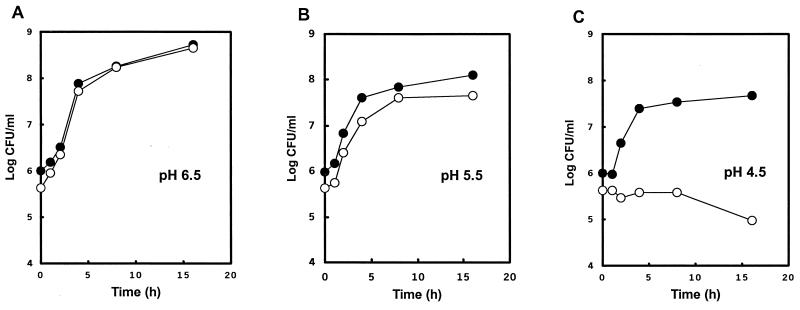
Although insertional mutagenesis abolished the function of wca, mutant M4020 at 37°C grew at a similar rate and survived as well as its isogenic parental strain W6-13. During acid treatment with pH levels ranging from 4.5 to 6.5, both W6-13 and M4020 grew in MGB at a similar rate at pH 6.5 (Fig. 2A); however, the growth of M4020 lagged slightly at pH 5.5 (Fig. 2B). The growth of W6-13 at pH 4.5 was not affected substantially compared to growth at pH 5.5 or 6.5; however, the growth of M4020 was completely inhibited at pH 4.5 (Fig. 2C). Similarly, M4020 was more susceptible than W6-13 to sublethal temperatures as reflected by significantly lower D-values at 55 and 60°C (Table 1).
FIG. 2.
Viability of E. coli O157:H7 W6-13 (●) and M4020 (○) at 37°C in MGB containing 0.5% Casamino Acids and adjusted to pH 4.5, 5.5, or 6.5 with 0.1 M HCl.
TABLE 1.
Effect of CA on thermal tolerance of E. coli O157:H7 wild-type strain W6-13 and its CA-deficient mutant M4020 in MGB containing 0.5% Casamino Acids
| Temp (°C) | Strain | Time (min) | Viable count (CFU/ml) | D-valuea (min) |
|---|---|---|---|---|
| 55 | W6-13 | 0 | 6.1 × 108 | |
| 15 | 1.3 × 108 | 26.4 | ||
| 60 | 3.0 × 106 | |||
| M4020 | 0 | 2.6 × 108 | ||
| 15 | 3.5 × 107 | 14.6 | ||
| 60 | 2.3 × 104 | |||
| 60 | W6-13 | 0 | 2.7 × 108 | |
| 5 | 1.3 × 105 | 1.67 | ||
| 10 | 2.8 × 102 | |||
| M4020 | 0 | 2.6 × 108 | ||
| 5 | 7.0 × 101 | 0.76 | ||
| 10 | <10 |
D-value, the time required to inactivate 90% of the bacterial population.
It has been suggested based on the results of previous studies that CA expressed on cell surfaces may simply act as a physical barrier to protect cells from hostile environmental conditions, such as enabling cells to retain water in a dry environment (1, 18, 21, 27). It was determined in another study that E. coli O157:H7 cells are rapidly inactivated by a rapid accumulation of protons (13). In an acidic environment, the lower the pH, the greater the concentration of protons. CA confers a strong negative charge to the cell surface which, we suggest may serve as a buffer by neutralizing protons at the cell surface, whereby preventing positively charged chemical groups from accumulating on cell envelopes and from penetrating into cells. We further hypothesize that the amount of CA on cell surfaces determines the buffering capacity of cells. When cells lose their ability to produce CA, cell surfaces become less negatively charged and thereby have reduced buffering capacity. When negatively charged cell surfaces are neutralized, protons will accumulate and enter cells freely. Such a change in intracellular pH will impair cell metabolism, causing cell death.
Studies revealed that the expression of CA is regulated by several direct and indirect mechanisms (9, 16, 19). The transcription of the wca operon is positively regulated by RcsA, RcsB, and RcsF and negatively regulated by ATP-dependent protease Lon (3, 11, 16, 29). RcsC can be activated by environmental stimuli and both positively and negatively regulates CA biosynthesis (29, 31). RcsC and RcsB act as a sensor and an effector, respectively, in a two-component regulatory system (9, 29, 31). Several heat shock proteins, including Dnak (Hsp70), GrpE, and DnaJ, also influence CA biosynthesis (16). A recent study revealed that DjlA, a member of the DnaJ family, is a DnaK cochaperone, and the induction of the colanic acid requires this DnaK-DjlA interaction to stimulate the RcsB-RcsC two-component signaling system in E. coli (7). Although CA on cell surfaces may act as a physical barrier and affect heat conductivity, thereby preventing heat from reaching cells readily, the influence of several heat shock proteins on the regulation of CA induction could further suggest a role for CA in the heat response of E. coli O157:H7.
Results of this study demonstrated that colanic acid is inducible in E. coli O157:H7, mostly by certain environmental stimuli, and that the induction of colanic acid has a substantial protective effect on the pathogen's acid and thermal tolerance. However, there are likely other factors of E. coli O157:H7 that confer to the pathogen additional protection against stressful environmental conditions. The mechanism of stress response in E. coli O157:H7 is complex, and other factors in addition to the induction of colanic acid are likely involved in conferring tolerance to the pathogen under stress-imposed conditions. Although there is limited information addressing the association of CA production with pathogenicity (1), a recent report suggests a role for colanic acid in the architecture of biofilms in the K-12 strain (6). Additional studies are under way to further elucidate the role of CA in the pathogen's exceptional tolerance to other forms of environmental stress, as well as in bacterial attachment and biofilm formation.
Acknowledgments
We thank Paul Danese in R. Kolter's lab, Harvard Medical School, for providing strain E. coli K-12 ZK2686.
This research was supported in part by a grant from the U.S. Department of Agriculture for the Alliance for Food Protection.
REFERENCES
- 1.Allen P M, Fisher D, Saunders J R, Hart C A. The role of capsular polysaccharide K21b of Klebsiella and of the structurally related colanic-acid polysaccharide of Escherichia coli in resistance to phagocytosis and serum killing. J Med Microbiol. 1987;24:363–370. doi: 10.1099/00222615-24-4-363. [DOI] [PubMed] [Google Scholar]
- 2.Benjamin M M, Datta A R. Acid tolerance of enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1995;61:1669–1672. doi: 10.1128/aem.61.4.1669-1672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brill J, Quinlan-Walshe A C, Gottesman S. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J Bacteriol. 1988;170:2599–2611. doi: 10.1128/jb.170.6.2599-2611.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Escherichia coli O157:H7 outbreak linked to commercially distributed dry-cured salami—Washington and California 1994. Morb Mortal Wkly Rep. 1995;44:157–160. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Outbreak of Escherichia coli O157:H7 infection and cryptosporidiosis associated with drinking unpasteurized apple cider—Connecticut and New York. Morb Mortal Wkly Rep. 1997;46:4–8. [PubMed] [Google Scholar]
- 6.Danese P N, Pratt L A, Kolter R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol. 2000;182:3593–3596. doi: 10.1128/jb.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genevaux P, Wawrzynow A, Zylicz M, Georgopoulos C, Kelly W L. DjlA is a third DnaK co-chaperone of Escherichia coli and DjlA-mediated induction of colanic acid capsule requires DjlA-DnaK interation. J Biol Chem. 2001;276:7906–7912. doi: 10.1074/jbc.M003855200. [DOI] [PubMed] [Google Scholar]
- 8.Goebel W F. Colanic acid. Proc Natl Acad Sci USA. 1963;49:464–471. doi: 10.1073/pnas.49.4.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman S, Stout V. Regulation of capsule polysaccharide synthesis in Escherichia coli K-12 biofilm architecture. J Bacteriol. 1991;182:3593–3596. [Google Scholar]
- 10.Grant W D, Sutherland I W, Wilkinson J F. Expopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae. J Bacteriol. 1969;100:1187–1193. doi: 10.1128/jb.100.3.1187-1193.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupte G, Woodward C, Stout V. Isolation and characterization of rcsB mutations that affect colanic acid capsule synthesis in Escherichia coli K-12. J Bacteriol. 1997;179:4328–4335. doi: 10.1128/jb.179.13.4328-4335.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jay J M. High-temperature food preservation and characteristics of thermophilic microorganisms. In: Jay J M, editor. Modern food microbiology. 4th ed. New York, N.Y: Van Nostrand Reinhold; 1992. p. 342. [Google Scholar]
- 13.Jordan K N, Oxford L, O'Byrne C P. Survival of low-pH stress by Escherichia coli O157:H7: correlation between alteration in the cell envelope and increased acid tolerance. Appl Environ Microbiol. 1999;65:3048–3055. doi: 10.1128/aem.65.7.3048-3055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Junkins A, Doyle M P. Demonstration of exopolysaccharide production by enterohemorrhagic Escherichia coli. Curr Microbiol. 1992;25:9–17. doi: 10.1007/BF01570076. [DOI] [PubMed] [Google Scholar]
- 15.Keene W E, Sazie E, Kok J, Rice D H, Hancock D D, Balan V K, Zhao T, Doyle M P. An outbreak of Escherichia coli O157:H7 infections traced to jerky made from deer meat. JAMA. 1997;15:1229–1231. doi: 10.1001/jama.1997.03540390059036. [DOI] [PubMed] [Google Scholar]
- 16.Kelly W L, Georgopoulos C. Positive control of the two-component RcsC/B signal transduction network by DjlA: a member of the DnaJ family of molecular chaperones in Escherichia coli. Mol Microbiol. 1997;25:913–931. doi: 10.1111/j.1365-2958.1997.mmi527.x. [DOI] [PubMed] [Google Scholar]
- 17.Leyer G J, Wang I, Johnson E A. Acid adaptation of Escherichia coli O157:H7 increases survival in acidic foods. Appl Environ Microbiol. 1995;61:3752–3755. doi: 10.1128/aem.61.10.3752-3755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Torres A J, Stout V. Role of colanic acid polysaccharide in serum resistance in vivo and in adherence. Curr Microbiol. 1996;33:383–389. doi: 10.1007/s002849900132. [DOI] [PubMed] [Google Scholar]
- 19.Markovitz A. Genetics and regulation of bacterial capsular polysaccharide biosynthesis and radiation sensitivity. In: Sutherland I W, editor. Surface carbohydrates of the prokaryotic cell. New York, N.Y: Academic Press, Inc.; 1977. pp. 415–462. [Google Scholar]
- 20.McClure P J, Hall S. Survival of Escherichia coli in foods. J Appl Microbiol. 2000;88:61S–70S. doi: 10.1111/j.1365-2672.2000.tb05333.x. [DOI] [PubMed] [Google Scholar]
- 21.Ophir T, Gutnick D L. A role of exopolysaccharides in the protection of microorganisms from desiccation. Appl Environ Microbiol. 1994;60:740–745. doi: 10.1128/aem.60.2.740-745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padhye N V, Doyle M P. Escherichia coli O157:H7: epidemiology, pathogenesis, and methods for detection in food. J Food Prot. 1993;55:555–565. doi: 10.4315/0362-028X-55.7.555. [DOI] [PubMed] [Google Scholar]
- 23.Rahn A, Drummelsmith J, Whitfield C. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J Bacteriol. 1999;181:2307–2313. doi: 10.1128/jb.181.7.2307-2313.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reitsma C J, Henning D R. Survival of enterohemorrhagic Escherichia coli O157:H7 during the manufacture and curing of Cheddar cheese. J Food Prot. 1996;59:460–468. doi: 10.4315/0362-028X-59.5.460. [DOI] [PubMed] [Google Scholar]
- 25.Riley L W, Remis R S, Helgerson S D, McGee H B, Wells J G, Davis B R, Herbert R J, Olcott E S, Johnson L M, Hargrett N T, Blake P A, Cohen M L. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 26.Riondan D C, Duffy G, Sheridan J J, Whiting R C, Blair I S, McDowell D A. Effects of acid adaptation, product pH, and heating on survival of Escherichia coli O157:H7 in pepperoni. Appl Environ Microbiol. 2000;66:1726–1729. doi: 10.1128/aem.66.4.1726-1729.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sledjeski D D, Gottesman S. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J Bacteriol. 1996;178:1204–1206. doi: 10.1128/jb.178.4.1204-1206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevenson G, Andrianopoulos K, Hobbs M, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torres-Cabassa A S, Gottesman S. Capsulre synthesis in Escherichia coli K-12 is regulated by proteolysis. J Bacteriol. 1987;169:981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wells J G, Davis B R, Wachsmuth L K, Riley L W, Remis R S, Sokolow R, Morris G K. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J Clin Microbiol. 1983;18:512–520. doi: 10.1128/jcm.18.3.512-520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitfield C, Valvano M A. Biosynthesis and expression of cell surface polysaccharides in gram-negative bacteria. Adv Microbiol Physiol. 1993;35:135–246. doi: 10.1016/s0065-2911(08)60099-5. [DOI] [PubMed] [Google Scholar]
- 32.Williams N C, Ingham S C. Thermotolerance of Escherichia coli O157:H7 ATCC 43894, Escherichia coli B, and an rpoS-deficient mutant of Escherichia coli O157:H7 ATCC 43895 following exposure to 1.5% acetic acid. J Food Prot. 1998;61:1184–1186. doi: 10.4315/0362-028x-61.9.1184. [DOI] [PubMed] [Google Scholar]