Entrez SNP (2.0) Updates:
May 26, 2020 - New Features....
1. Search dbSNP using HGVS**
Example: NM_000237.3:c.1421C>G OR NG_013007.1:g.7147G>A
2. SNPs with ALFA frequency**
All SNPs with ALFA frequency can be retrieved using an Entrez filter.
Example: all[sb] AND by alfa[Validation]
User 'by-ALFA' facet under 'Validate Status' Filters.

Click on the 'ALFA' link of each RefSNP to view frequency by population.
February 5, 2020 - Support for GRCh37 position search and reporting
dbSNP web search (https://www.ncbi.nlm.nih.gov/snp/) and eUtils eSearch, eFetch, and eSummary now supports both GRCh38 and GRCh37(new) coordinates. The POSITION_GRCH37 field have been added to the guide of all available terms.
Example: Search rs1000000038 using the GRCh38 and GRCh37 fields POSITION and POSITION_GRCH37, respectively
GRCh38 -
https://www.ncbi.nlm.nih.gov/snp/?term=62587167%5BPOSITION%5D+AND+8%5BCHR%5D
GRCh37 -
https://www.ncbi.nlm.nih.gov/snp/?term=63499726%5BPOSITION_GRCH37%5D+AND+8%5BCHR%5D
dbSNP eUtils eFetch and eSummary XML report GRCh37 positions in the previous assembly field CHRPOS_PREV_ASSM along with chromosome positions (CHRPOS) the variant positions on the latest assembly (GRCh38).
Example: https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=snp&retmode=xml&id=328
GRCh38:
<CHRPOS>8:19962213</CHRPOS>
GRCh37:
<CHRPOS_PREV_ASSM>8:19819724</CHRPOS_PREV_ASSM>
Visit the resources below for additional information.
Search guide - https://www.ncbi.nlm.nih.gov/snp/docs/entrez_help/
eUtils tutorials - https://github.com/ncbi/dbsnp/tree/master/tutorials
Please send your comments and suggestions to ([email protected]).
September 29, 2019 - dbSNP 2.0 General Updates
Notifications: As previously announced on April 19, 2018, dbSNP Entrez currently only houses human data. In addition, the Entrez and eUtils report formats RS docsum, XML, ASN.1, FASTA, and FLAT format will no longer be available. dbSNP Entrez eUtils will transition to a new single compact eSummary format, the spec and sample data are below. Please contact [email protected] if you have any comments or concerns.
No change to the web search https://www.ncbi.nlm.nih.gov/snp
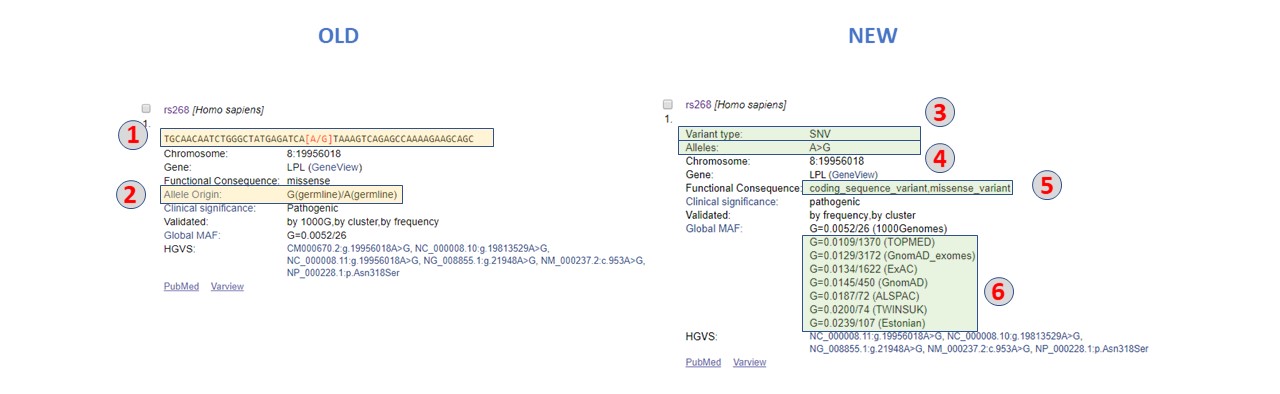
Web Result Display changes
1. Removed flanking sequences
2. Removed Allele Origin
3. Removed Geneview
4. Added Variant Type - Variant types supported.
5. Added Alleles - The alleles are always reported in the forward orientation with respect to the latest assembly version (ie. GRCh38.p13).
6. Added additional Functional Consequences - All consequences based on RefSeq are reported, not just most severe as reported in old version.
7. Added additional Population Frequency - Allele frequency from different projects are added in addition to the previously reported Global MAF that was based on the 1000 Genomes Project.
Eutils changes
1. Deprecated eUtils reports - RS docsum, XML, ASN.1, FASTA, and FLAT are deprecated and no longer supported.
2. Added eUtils RefSNP JSON report (beta) - Use efetch to retrieve dbSNP new JSON object (schema) containing comprehensive RefSNP content and for convenient integration into eUtils workflows. This beta service is still in testing mode so please limit your request to 1-3 RSID/second. Alternative, you can use the Variation Service API directly with additional functions including variant normalization using RSID, SPDI, HGVS, or VCF format and remapping. Complete downloads of dbSNP (+660M RS) are also available on the FTP site in JSON or VCF formats.
efetch example:
curl -X GET "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=snp&id=268,328&retmode=text&rettype=json"
3. Updated eSummary XML - The single XML eSummary report is available with the changes listed (1-5) above. A sample report and schema are available on GitHub.
eSummary example: https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esummary.fcgi?db=snp&id=268,328
curl -X GET "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esummary.fcgi?db=snp&id=268,328"
Users should migrate to the new default eSummary 2.0 version above. The older 1.0 version will be deprecated later this year but is still made available during the transition period. The old 1.0 version can be requested by adding the version number (version=1.0) as shown below. https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esummary.fcgi?db=snp&id=268,328&version=1.0
dbSNP Tutorials
Tutorial are available on GitHub.
