Searching dbSNP in Entrez
Table of contents
How to construct queries
dbSNP is part of NCBI's network of Entrez databases. As with these other databases, data of interest may be located simply by entering keywords into the dbSNP search box. The Advanced Search page, linked below the dbSNP search box, can assist in the construction of complex queries. To construct a complex query, specify the search terms, their fields, and the Boolean operations to perform on the terms using the following syntax:
term[field] OPERATOR term[field]
where term is the search terms, field is the search field, and OPERATOR is the Boolean operator ('AND', 'OR', 'NOT'; must be capitalized).
Common Query fields and examples
| Field full name | Field aliases | Description | Search term values and rules | Example |
|---|---|---|---|---|
| All Fields | ALL, * | Search all searchable (indexed) fields | Asterisk (*) in the search term is not interpreted as a wildcard | SNV AND pathogenic |
| Base Position | POSITION, SNPPOS | Chromosome base position on GRCh38 (current) | A natural number representing the SNP's start coordinate on its chromosome on the latest assembly (ie. GRCh38). Most useful when search in combination with the CHR field. | 19956018[POSITION] AND 8[CHR] |
| Base Position Previous | POSITION_GRCH37, CHRPOS_PREV_ASSM | Chromosome base position on GRCh37 (previous) | A natural number representing the SNP's start coordinate on its chromosome on the previous assembly (ie. GRCh37). Most useful when search in combination with the CHR field. | 19813529[POSITION_GRCH37] AND 8[CHR] |
| Chromosome | CHR, CHRNUM | Chromosomes | One of 1-22, X, Y, MT | 7[CHR] |
| Clinical Significance | CLIN | Variations with defined clinical effects or significances | 16 search term values, defined for a relatively small subset of SNPs. | "likely pathogenic"[CLIN] |
| Filter | FILT, FLTR, SUBSET, SB, FIL | Limits the records returned | A variety of filters is available, including functional, positional, source, etc. | get all dbSNP records "all[sb]" or subsets "splice 5 snp"[Filter] |
| Function Class | FXN, Function_class, FUNC, FUNCTION, FUNCTION_CLASS | Function class | 21 function classes are defined | "frameshift"[Function Class] |
| Gene Name | GENE, GENE_SYMBOL | Entrez Gene symbol | Corresponds to the Official Symbol field in the Entrez Gene resource | MAPK1[GENE] |
| Gene ID | GENE_ID | Entrez Gene UID | The numeric ID referencing the Entrez Gene ID | 5594[GENE_ID] |
| Global Minor Allele Frequency | GMAF | Minor Allele Frequency derived from global population (i.e., 1000G); can also be study-wide MAF that is not from global population | Most useful when entered as a range, as in the example | (0.0[GMAF] : 0.01[GMAF]) |
| Project or Submitter Handle | HAN, PROJECT | Submitter Handle or Project Name | Submitter lab or project name including 1000Genomes, GnomAD, and DebNick | 1000genomes[Submitter Handle] or 1000genomes[PROJECT] |
| Reference SNP ID | RS, SNPID | Clustered SNP ID (rs) | The numeric ID must be prefixed with "rs". Also retrieves SNPs that have been merged into the specified SNP. | rs328[RS] |
| SNP Class | SCLS, SNPCLASS | SNP class | Possible values are: "del", "delins", "ins", "mnv", and "snv". | del[SNPCLASS] |
| Submitter SNP ID | SS, SSNUM | The ID assigned to each report of a SNP at submission time | Must be prefixed with "ss". Note that the query still returns Reference SNPs rather than Submitter SNPs. | ss329[SS] |
| Validation Status | VALI, VALIDATE, VALIDATION | Validation status | Possible values are: "by cluster" or "by frequency" | "by cluster"[Validation Status] |
Complex queries and others
| Description | Query | Note |
|---|---|---|
| Variant allele with MAF = 0 | "00000.0000"[Global Minor Allele Frequency] | variant allele is homozygous and may be due to differences between assembly versions |
| Pathogenic variants in BRCA1 with MAF < 0.01 | "pathogenic"[Clinical Significance] AND BRCA1 AND 00000.0000:00000.00999[GLOBAL_MAF] | set GLOBAL_MAF range between 0 and 0.00999 for MAF < 0.01 |
| Common variant (MAF => 0.01) | 00000.0100[GLOBAL_MAF] : 00001.0000[GLOBAL_MAF] | set GLOBAL_MAF range from 0.0100 and 1.0000 for MAF => 0.01 |
| 1000Genomes common variant (MAF => 0.01) not found by TOPMED | "1000genomes"[Submitter Handle] NOT "topmed"[Submitter Handle] AND 00000.0100: 00001.0000[GLOBAL_MAF] | set GLOBAL_MAF range from 0.0100 and 1.0000 for MAF => 0.01 |
HGVS Search
Users can search dbSNP with HGVS names, as shown with the example: NM_000237.3:c.1421C>G OR NG_013007.1:g.7147G>A

ALFA Frequency
All SNPs with ALFA frequency
To retrieve the complete list of variations with ALFA frequency information, we can use this query: all[sb] AND by alfa[Validation]
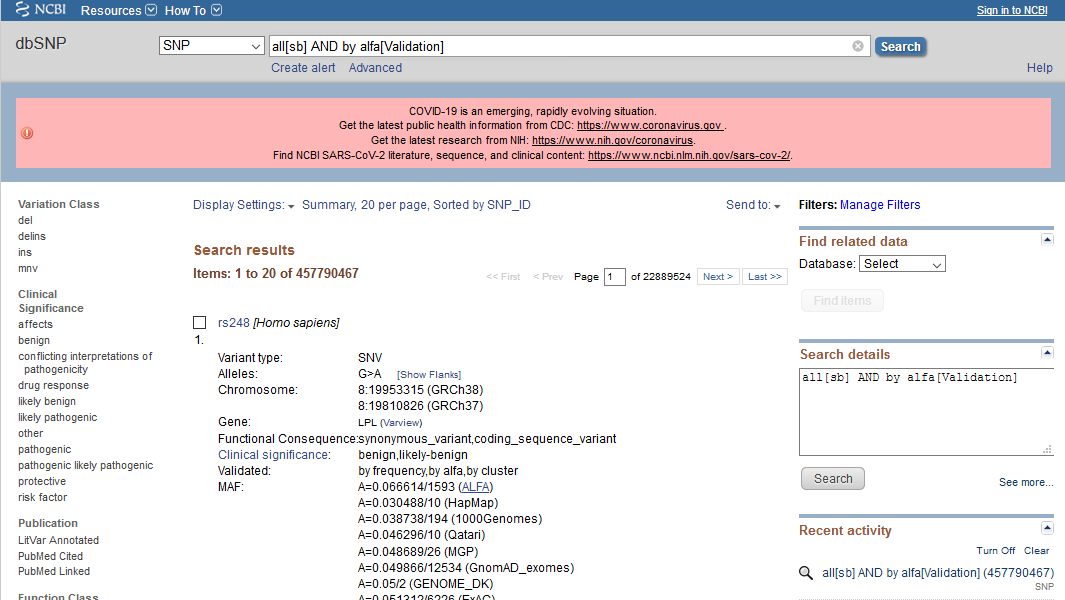
'by-ALFA' Facet
There is a 'by-ALFA' facet under 'Validate Status' filters, which can be used to further filter out the search result.

'ALFA' Link
For each RefSNP in the search result, there is an 'ALFA' link that leads to frequency by population.
Protein Position
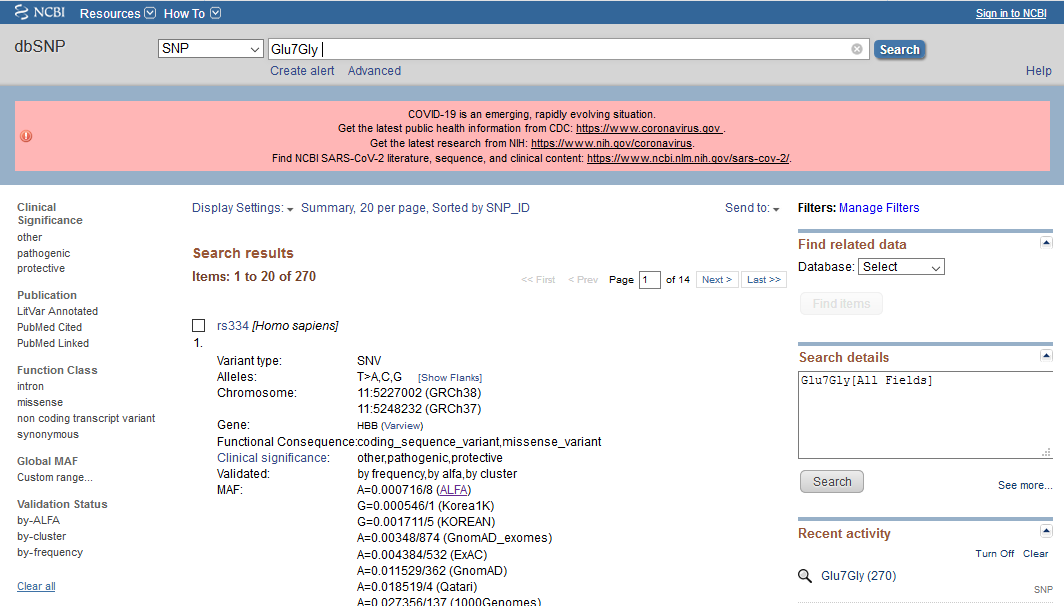
It's also possible to search by amino acid variation at a protein sequence level. The image below shows the result for a search string 'Glu7Gly'.