ALFA: Allele Frequency Aggregator
Table of Contents
ALFA at a glance:
- The goal is to make allele frequency data from over 1 million subjects available in dbGaP as open-access in accordance with the FAIR Data Principles (Findable, Accessible, Interoperable, and Reusable).
- The dbGaP studies include chip array, exome, and genomic sequencing data with subjects from 12 diverse populations including European, African, Asian, Latin American, and others.
- The data will be integrated with dbSNP regular build release with assigned RS accessions for variants and available for access by web, FTP, API, and TrackHub.
Background
The NCBI database of Genotypes and Phenotypes (dbGaP) contains the findings of over 2K studies on the interaction of genotype and phenotype. The database has over three million subjects and hundreds of millions of variants along with thousands of phenotypes and molecular assay data. This unprecedented volume and diversity of data offers enormous potential for identifying genetic factors that influence health and disease. The National Institutes of Health (NIH) recently has lifted the restriction on Genomic Summary Results (GSR) access for responsible data sharing and use.
In order to comply with the updated GSR policy and to encourage research aimed at identifying genetic variants that contribute to health and disease, NCBI created the Allele Frequency Aggregator (ALFA) pipeline, which computes allele frequency for variants in dbGaP across approved unrestricted studies and makes the data available to the public via dbSNP. The ALFA project's goal is to make frequency data from over 1M dbGaP subjects open-access to aid in the discovery and interpretation of common and rare variants with biological implications or causing diseases. Almost ~1M subjects with genotype data have been analyzed using GRAF-pop as ALFA project candidates, pending study approval and processing.
Build Summary
| Release | Version | Date |
|---|---|---|
| 1 | 20200227123210 | March 10, 2020 |
| 2 | 20201027095038 | January 6, 2021 |
| 3 | 20230706150541 | August 2, 2023 |
Data Generation
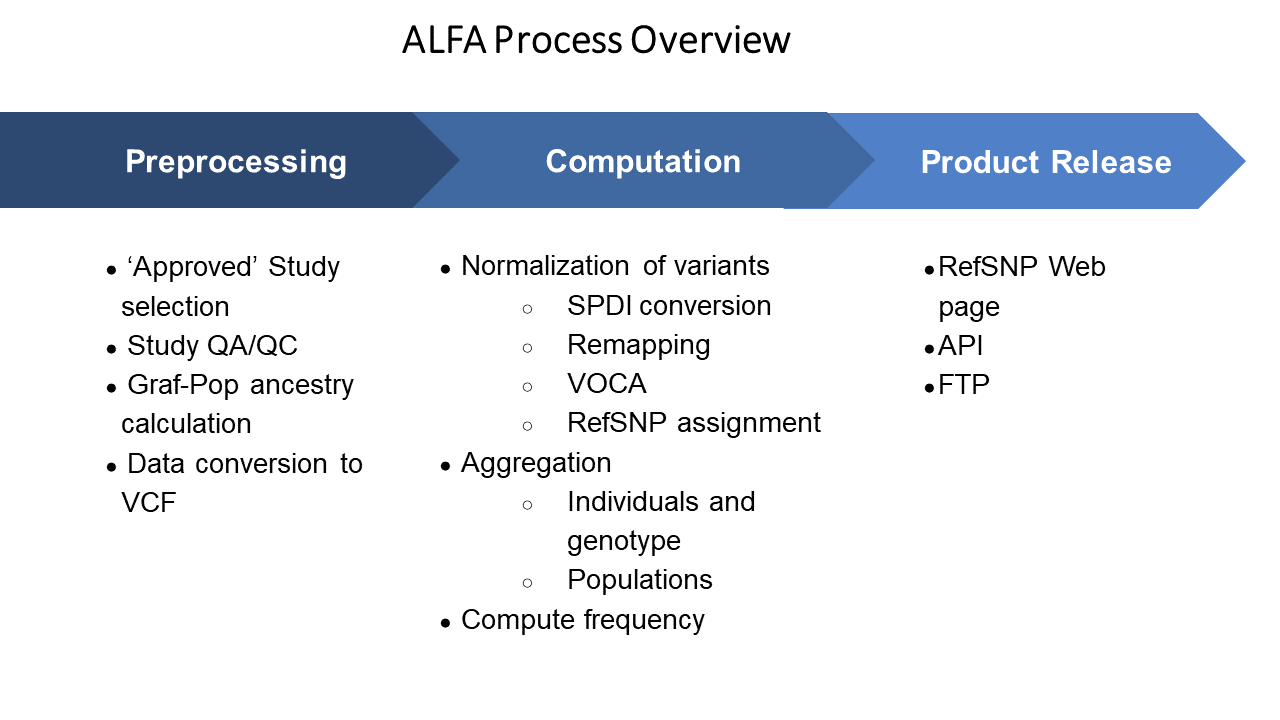
Data from selected studies are harmonized and normalized. Using existing dbSNP and dbGaP curation and semi-automatic pipelines the data either from GWAS chip array genotyping or direct sequencing of exomes and whole genomes were QA/QC and transformed to standard VCF format as input into a pipeline that transform variants to SPDI notation and normalized using VOCA to aggregate, remap and cluster to existing dbSNP rs or assign new ones (Holmes et al.), and allele frequency computed.
Populations
Sample ancestries are validated using GRAF-pop (Updated Sept 2021) and assigned to 12 major populations including European, Hispanic, African, Asian, and others (Jin et al., 2019).
Data QC
We do our best to ensure that the data released is of the highest quality, complete, accurate, and useful. However, because we did not generate the original submitted data from dbGaP that were used as input for this project, and because the processing required to make the data useful is complex, we cannot be liable for omissions or inaccuracies. Please see the release summary with QC report (coming soon) for more details.
Data Excluded by QC:
-
Variants with call rate < 95%
-
Subjects with call rate < 95%
Data Excluded by QC and awaiting fixes from original dbGaP Submitters and may be included in future releases.
-
Array datasets with conflicting subjects or markers between the marker manifest and reported genotype
-
Datasets with incorrect or flipped allele orientation
-
Datasets where the frequency of Ancestry Informative Markers (AIMs) tested is inconsistent with 1000 Genomes for whole study or for a particular population. The dataset is excluded if the percentage of AIMs outlier markers tested with allele frequency difference > abs(+/-0.15) exceed 0.3% for the whole study or 0.1% for a population (see details).
-
Dataset where polymorphic SNPs are recorded as monomorphic
-
Dataset suspected of having errors due to chip array design
-
Dataset with various systemic errors and not does not appeared random
Terms of Use
Please see the Terms of Use applied to dbGaP frequency data and NCBI standard disclaimer.
Data Release Cycle
ALFA import new studies and regenerate the data in a quarterly basis for release with each dbSNP build. We anticipate adding between 100-200K new dbGaP subjects per release. Novel variants will be assigned RS numbers and the frequency data will be integrated with dbSNP regular release products (Entrez search, RefSNP report, API, Sequence Viewer, Variation Viewer, and FTP JSON and VCF files).
Interim ALFA releases to provide more frequent updates, such as the initial release, will only include reporting of ALFA allele frequency for existing RS on the RefSNP page. Separate ALFA specific download files are provided that include both existing RS and novel variants. Novel variants from interim releases are also available by API position search (See Data Access below).
Users can subscribe to the mailing list to get data release and update announcements.
Data Access
RefSNP Web Page
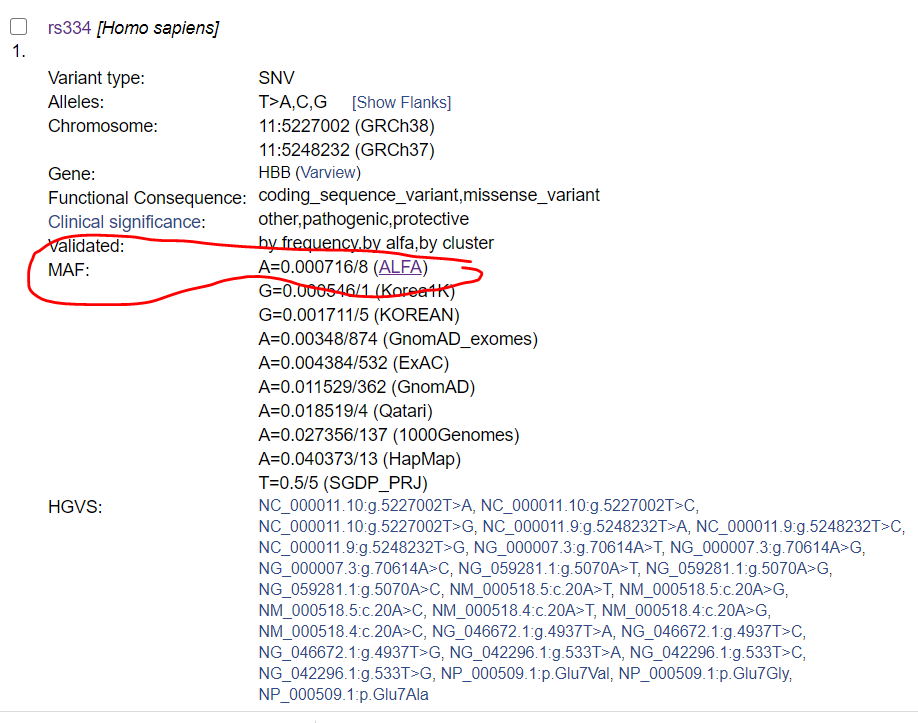
Access RefSNP page using the rs number. Allele frequencies from ALFA and other projects are reported in "Frequency" tab.
Example: rs334
FTP Download
All ALFA dbGaP variants including novel ones not yet in dbSNP are available in VCF format.
- Directory: https://ftp.ncbi.nih.gov/snp/population_frequency/
- Variants reported on GRCh38. Please see the Readme for more details and exceptions.
Track Hub
An ALFA track hub definition file can be used to add ALFA tracks to a personalized Genome Browser or Genome Data Viewer. An example showing this hub in NCBI's graphical viewer is here.
An ALFA track hub is also now publicly available with the UCSC Genome Browser. It can be acceessed with this link.
API Queries
All ALFA dbGaP variants including novel ones not yet in dbSNP are available through NCBI Variation Service API and include three queries:
-
By Position using GRCh38 coordinates
-
By RefSNP(rs) using dbSNP accession.
See tutorials below for Python examples.
Enhanced Search and Filtering Features
More search and filtering features are added to NCBI search page to make use of the ALFA frequency data.
ALFA Reporting on Entrez SNP
On the SNP search result page, if an RS has ALFA frequency information, it will be displayed along with a url link to the frequency tab on the SNP RS page.
Search Filtering with ALFA
A user can also filter the search results with ALFA frequency. As shown in the image below, on the left side of the search result page, a filter 'by-ALFA' is added under Validation Status.

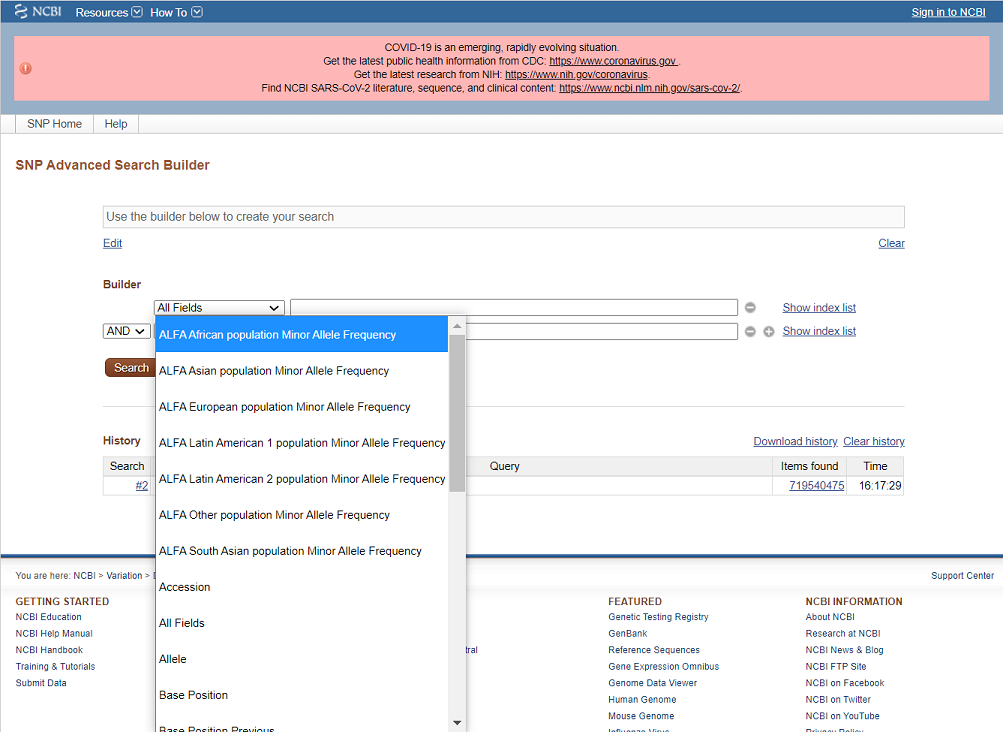
Advanced Search with ALFA Population
With the SNP Advanced Search Builder, a user can search RS with ALFA frequency of a specific population. The user first selects a population from the dropdown list and then provides a specific range of the minor allele frequency.
Tutorials
-
Gene
-
Locations
-
Additional Variation Services API Tutorials are available here.
Presentations
- NCBI Minute: ALFA Webinar materials and video.
- ASHG 2019 Collab
- ASHG 2019 Platform talk
- Human Population Genetic Data at NCBI (Video)
- New Variation Services for Normalizing, Remapping, and Annotating Variants (Video)
- ASHG 2020 Collab: Introduction to and tutorial for using ALFA, the Allele Frequency Aggregator, at the National Center for Biotechnology Information (NCBI) (Video).
Citing this Project
We're planning on submitting a resource manuscript about the ALFA project later this year. For now, please use the MLA standards for citing this project website below.
L. Phan, Y. Jin, H. Zhang, W. Qiang, E. Shekhtman, D. Shao, D. Revoe, R. Villamarin, E. Ivanchenko, M. Kimura, Z. Y. Wang, L. Hao, N. Sharopova, M. Bihan, A. Sturcke, M. Lee, N. Popova, W. Wu, C. Bastiani, M. Ward, J. B. Holmes, V. Lyoshin, K. Kaur, E. Moyer, M. Feolo, and B. L. Kattman. "ALFA: Allele Frequency Aggregator." National Center for Biotechnology Information, U.S. National Library of Medicine, 10 Mar. 2020, www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa/.
Contact
Please send your comments and suggestions to [email protected]
