To access SPARCLE, you can either:
With either approach, the corresponding SPARCLE record(s) will display the name and functional label of the protein's conserved domain architecture, supporting evidence, and links to other proteins with the same architecture. Details about each approach are below.
 
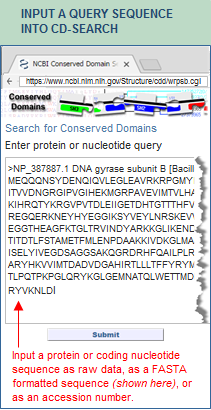
- The most common way to access SPARCLE is to enter a query sequence into CD-Search, either as FASTA-formatted sequence data, or as an accession number of a sequence that is in the protein or nucleotide databases. The search results will include a "Protein Classification" section if the query protein has a hit to a curated domain architecture in the SPARCLE database. In the protein classification section, click on the domain architecture ID in order to open the corresponding SPARCLE record.
- The illustration below provides an example, using NP_387887, DNA gyrase subunit B, as the protein query sequence.
- You can click on the individual panels of the illustration to open the corresponding live web page:
-
Ongoing research: The Conserved Domain Database (CDD), as well as the conserved domain architecture annotated on proteins by SPARCLE, continue to evolve as new data become available and as research progresses. Therefore, the live web page views might differ from the illustration above.
For example, in January 2017, the protein sequence NP_387887 was initially annotated with architecture ID 10647733 (as shown in the illustration above). That architecture is named "DNA gyrase subunit B" and includes four distinct conserved domains.
In March 2017, when a new build of CDD/SPARCLE was released, the conserved domain architecture annotation for NP_387887 was revised to architecture ID 11481348 (which is a multi-domain that encompasses the four original conserved domains, and which can be seen in the current CD-Search results for NP_387887). That architecture has a more specific and precise name, "type IIA DNA topoisomerase subunit B," and reflects the full length protein model.
To see the four distinct conserved domains that compose the full length protein model, simply change the CD-Search display option on the live CD-Search results for NP_387887 from "Concise Results" to "Full Results" (using the "View" menu near the upper right hand corner of the CD-Search results page). The Full Results display will show the four conserved domains that compose the full length protein model.
As the available data and understanding of conserved domain architectures continue to evolve, the domain architectures that are annotated on proteins may evolve as well, as shown in this example. Comments about the data are welcome and can be sent to the NCBI Support Center/Help Desk, which is accessible as a link in the footer of NCBI web pages.
- Additional details about using the CD-Search tool are provided in the CD-Search Help Document.
 
- The SPARCLE database can be searched by keyword. That will retrieve domain architectures that contain the term(s) of interest in their descriptions.
- The illustration below provides an example. It searches the SPARCLE database for conserved domain architecture records that contain the terms "chloride" and "channel", and limits the results to curated domain architecture records by adding curated[ReviewLevel] to the search.
- Click on the individual panels of the illustration below to open the corresponding live web page:
- Beneath the illustration are additional details about:
- the scope of a search, describing which fields of a database record are searched
- search tips for narrowing or broadening your search:
- a tabular list of search fields, including a description and sample search for each field
- Scope of a keyword search:
 
- When you search the SPARCLE database by keyword (e.g., gyrase), All Fields are searched by default. This includes looking for your keyword(s) in the name & functional label (description) of the conserved domain architecture. This also includes looking for your keyword(s) in the entities that were used as evidence to give a name to the architecture, such as gene names (names of genes whose protein products have that architecture), protein names (definition lines of proteins used as evidence to support the domain architecture, such as SwissProt records, where protein sequences are named based on literature), conserved domain names (including the short and long names of conserved domains that are present in the architecture), Enzyme Commission (EC) numbers and corresponding EC text descriptions.
- Search tips for keyword searches:
 
all fields are searched by default | how to limit your query to a specific search field |
use quotes to force a phrase search |
use an asterisk (*) for truncation |
compare some sample search strategies
- By default, All Fields are searched in the SPARCLE database.
- Limit a query to a specific search field:
If you prefer to narrow your search to a specific field, you can:
- Use the "Limits" page or the "Advanced" search page to view a list of available search fields, and select the field of interest from a pull-down menu.
- Alternatively, you can type the field name, surrounced by square brackets [], directly after your search term, with or without a space between your term and the first bracket. For example:
- a search for: curated[ReviewLevel] looks for the term "curated" in the "ReviewLevel" search field
- a search for: bacteria[Organism] looks for the term "bacteria" only in the "Organism" search field. This will retrieve conserved domain architectures whose names and labels are applicable within bacteria but not within other taxonomic nodes.
- The available search fields are listed in a table below, including a description and search example for each field.
- A footnote under the table shows how search fields can be specified using either their full spelling or an abbreviation, and in upper case, lower case, or mixed case.
- The "Show Index" link on the SPARCLE Advanced Search page allows you to browse the index of each search field, where you can see the available terms, the number of records containing each term or phrase, as well as the syntax for entering values in search fields such as CreateDate.
- Use quotes to search for a phrase:
Another way to narrow your search is to enclose multiple terms in quotes (e.g., search for "chloride channel").
- Using quotes will require the system to search for the terms as a phrase. It will therefore only retrieve records where the two words occur together, adjacent to each other.
- If quotes are not used, the Entrez system may still recognize and handle the terms as a phrase, if they are present in a phrase dictionary used by the search engine. If the terms are not present in the phrase dictionary and are not surrounded by quotes, Entrez will insert a Boolean AND between the terms; in that case, they may or may not appear adjacent to each other in the retrieved records.
- The "Details" section in the right hand margin of a search results page will show you exactly how the Entrez system parsed your query. More search tips are provided in the PubMed help document and Entrez help document.
- Use an asterisk (*) for truncation
To broaden a search, you can use an asterisk (*) as a wild card to search for a word stem.
- For example, a search for chlori* will retrieve records with terms such as chloride, chlorin, chlorinate, chlorinated, chlorinating, chlorination, chlorine, chlorite, and chloritidismutans.
- As another example, a search for arachidon* will retrieve records with terms such as arachidonate, arachidonic, arachidonoyl, and arachidonyl.
- The Entrez Help document provides additional information about truncating search terms in this way.
- Compare some sample search strategies:
As examples of various search strategies, compare the results of the following searches:
- chlori*
If an asterisk is used to truncate a search term, the system will retrieve all records that contain the specified word stem. The word stem can appear in any field of the record, unless you specify a desired search field.
- chloride channel
If no search field is specified, [All Fields] are searched by default, Also, the keywords are not necessarily searched as a phrase, but can occur separately in different parts of the record.
- "chloride channel"
Use quotes to search for the terms as a phrase.
- "chloride channel"[Name]
Limit the query to a specific search field, such as the [Name] field shown here, to narrow the search results.
- "chloride channel" AND curated[ReviewLevel]
Add a [ReviewLevel] criterion to the query, as shown above, to limit retrieval to a specified subset of architectures (e.g., architectures that have been curated, autonamed, or namedByDomain).
- Search Fields:
 
As noted in the Search Tips above, when you search the SPARCLE database by keyword, All Fields are searched by default. If you prefer to restrict your search to a specific data field, you can use the pull-down menus on either the "Limits or the "Advanced" search page to select the desired field. Alternatively, you can type the desired field directly in your query, surrounding field name with square brackets [].*
| |
| Field name |
Abbreviation* |
Description |
Sample Search |
| All Fields |
[All]
[All Fields] |
Searches all of the indexed fields in the SPARCLE database.
If no field specifier is included in a query, the system searches [All] fields by default, as happens with the first sample search shown at the right. Click on that search to open the corresponding results page. The "Search Details" box that appears in the right hand margin of the search results page shows that the query was translated by the system to:
chloride[All Fields] AND channel[All Fields]
|
 chloride channel
chloride channel
The basic search above, in which the query terms are entered without quotes, will retrieve the architecture(s) that contain the word "chloride" and the word "channel" in any field of the record. The words do not have to be adjacent to each other in the record (i.e., they do not have to appear as a phrase), and they do not have to appear in the same field.
"chloride channel"[all]
The search above, which surrounds the search terms with quotes, will retrieve the architecture(s) that contain the phrase "chloride channel" in any field of the record. (The quotes surrounding the search terms ensure they are searched as a phrase.)
Note: Compare the results of the above search, which looks for the phrase "chloride channel" in any field of the record, with the more specific results obtained by the sample [Name] field search:
"chloride channel"[Name]
which retrieves records containing the phrase "chloride channel" only in the name of the conserved domain architecture.
(The data processing section of this document describes how architectures are named.)
|
| BiosystemsDescription |
[BiosystemsDescription] |
Descriptions of BioSystems that are listed as supporting evidence for conserved domain architectures in the SPARCLE database.
As noted on the About BioSystems page, a biosystem is a group of molecules that interact in a biological system. One type of biosystem is a biological pathway, which can consist of interacting genes, proteins, and small molecules. Another type of biosystem is a disease, which can involve components such as genes, biomarkers, and drugs.
|
 "folate biosynthesis"[BiosystemsDescription]
"folate biosynthesis"[BiosystemsDescription]
will retrieve architecture(s) that list, as supporting evidence, biosystems whose descriptions contain the phrase "folate biosynthesis."
|
| CDDDescription |
[CDDDescription] |
Description of conserved domain models that are components of, or that are listed as supporting evidence for, conserved domain architectures in the SPARCLE database. |
 "transport proteins"[CDDDescription]
"transport proteins"[CDDDescription]
will retrieve architecture(s) that contain conserved domain models whose description includes the phrase "transport proteins."
|
| CDDShortname |
[CDDShortname] |
Short names of conserved domain models that are components of, or that are listed as supporting evidence for, conserved domain architectures in the SPARCLE database.
The short name is the label that appears on the conserved domain's cartoon in a CD-Search results display.
Note: This field can only be searched by entering the complete short name, surrounded by quotes. Entering a single term or other fragment from the short name will not retrieve results. (See examples below.)
Because of this, it is better to search the [CDDDescription] field because it offers more comprehensive searches.
--------------------
Examples: To illustrate the use of the [CDDShortname] field:
A search for the following complete string: "voltage gated clc"[CDDShortname] will retrieve architectures that contain a conserved domain model with that short name.
However, a search for the single word: voltage[CDDShortname] will not retrieve any records, because there are no conserved domains that have a short title of the single word "voltage."
--------------------
Tip: The Advanced search page can be used to browse the available terms in any index.
For example, to see a list of short names, use the "Builder" section of the advanced search page, select the CDDShortname search field from the pull-down menu, then click on "Show index list."
Note: If you do not enter any term in the text box beside the selected search field, the system will automatically take you to the top of the index for the selected search field, and you can then scroll through the terms.
If you enter a term in the text box before clicking on "Show index list," the search system will jump to the part of the index that contains your term, then you can scroll up or down.
|
 "voltage gated clc"[CDDShortname]
"voltage gated clc"[CDDShortname]
will retrieve architecture(s) that contain a conserved domain model whose short name is "voltage gated clc".
(The quotes surrounding the search terms ensure they are searched as a phrase.)
|
| CDDTitle |
[CDDTitle] |
Title of conserved domain models that are components of, or that are listed as supporting evidence for, conserved domain architectures in the SPARCLE database.
Note: Some older conserved domain models do not have a title. For example, the conserved domain model with accession cd00400 has a short name of "Voltage_gated_ClC" and an extensive description, but it doesn't have a separate title. As a result, those records will not be retrieved by a search of the [CDDTitle] field.
Therefore, is generally better to search for the [CDDDescription] field, rather than the [CDDTitle] field, because the [CDDDescription] field provides a more comprehensive search.
For example, compare the results of the [CDDTitle] and [CDDDescription] searches:
voltage[CDDTitle]
vs.
voltage[CDDDescription]
|
 voltage[CDDTitle]
voltage[CDDTitle]
will retrieve architecture(s) that contain a conserved domain model whose title includes the word "voltage".
"voltage gated chloride channel"[CDDTitle]
will retrieve architecture(s) that contain a conserved domain model whose title includes the phrase "voltage gated chloride channel".
Note: It is generally better to search for the [CDDDescription] field, rather than the [CDDTitle] field, because the [CDDDescription] field provides a more comprehensive search. See the note and examples in the preceding column.
|
| Comment |
[Comment] |
The [Comment] field contains free text that was written by curators in the supporting evidence fields of SPARCLE records. It represents something the curators wanted to note about the conserved domain architecture, based on the research they did in curating and naming the architecture.
|
 chloride[Comment]
chloride[Comment]
will retrieve the architectures that contain the word "chloride" in the comments section of a conserved domain architecture's supporting evidence.
|
| CreateDate |
[CreateDate]
[CDAT]
[PDAT]
[DP] |
The date on which the current version of a conserved domain architecture record was published in the SPARCLE curation system.
This is referred to as the Create Date [CDAT]. Alternatively, it is sometimes referred to as the Publication Date, or Date of Publication, hence the alternative abbreviations of [PDAT] or [DP].
The architecture subsequently becomes available in the public SPARCLE database, although that might happen a bit later.
Examples:
--------------------
To search for a specific day, month, or year, enter it in any one of the following formats:
YYYY/MM/DD
will retrieve all architectures that were published in the SPARCLE curation system on the specified day
or
YYYY/MM
will retrieve all architectures that were published in the SPARCLE curation system in the specified month
or
YYYY
will retrieve all architectures that were published in the SPARCLE curation system in the specified year
--------------------
To search for a range of dates, enter your in any one of the following formats, using the colon (:) as the range operator:
YYYY/MM/DD[CDAT]:YYYY/MM/DD[CDAT]
will retrieve all architectures that were published in the SPARCLE curation system between the two dates you specified
|
 Single date:
Single date:
2017/04/20[CDAT]
will retrieve all architectures that were published in the SPARCLE curation system on 20 April 2017.
Date range:
2017/04/20[CreateDate] : 2017/05/18[CreateDate]
will retrieve all architectures that were published in the SPARCLE curation system between 20 April 2017 and 18 May 2017.
In the query above, the colon (:) serves as the range operator.
|
| Defline |
[Defline] |
The definition line (description) of any protein sequence that was used as supporting evidence for a conserved domain architecture. |
 chloride[defline]
chloride[defline]
will retrieve the architectures that list, as supporting evidence, any proteins that have the term "chloride" in their definition line.
|
| ECNumber |
[ECNumber] |
The Enzyme Commission (EC) number that is found in the sequence record of any protein that was used as evidence for a conserved domain architecture, or the EC number that is found in a high quality (e.g., curated) sequence record that belongs to the group of proteins annotated with the architecture.
The Enzyme Nomenclature and Classification system is based on the reactions catalyzed by the enzymes. The system is developed by one of the Nomenclature Committees of the International Union of Biochemistry and Molecular Biology (IUBMB). Separate websites enable you to browse enzymes by class, or to search the enzyme nomenclature database by text word or number.
--------------------
Method for assigning EC numbers to conserved domain architecture records in SPARCLE:
Typically, the EC numbers are taken from Swiss-Prot records that belong to the cluster of proteins that have a given architecture.
In addition, the EC number from a Swiss-Prot record might also be applied to other, similar protein clusters that essentially represent the same architecture. Those architectures might have been split into separate SPARCLE records only because they contain slightly different domain models. For example, two or more protein clusters might have top-scoring hits to overalapping/redundant conserved domain models from different source databases, but their architectures are essentially similar, as in the hypothetical example below.
--------------------
As a hypothetical example of how an EC Number from one architecture might be annotated on other architectures:
a) Let's say you have three architectures that are similar to each other:
- They each have their own SPARCLE record because their top scoring domain models are slightly different from each other:
- ------[pfam01]------[pfam05]------
- ------[pfam01]------[COG12]------
- ------[pfam01]------[cd0008]------
b) Let's also say that:
- domain models pfam05, COG12, and cd0008 are redundant (i.e., they come from different source databases, but they overlap with each other on protein sequences and are therefore redundant)
- architecture #2 maps to protein sequence SwissProt P0321
- SwissProt P0321 has been annotated with an EC number.
c) As a result:
- architectures #1, 2, and 3 above are essentially the same (due to the redundant nature of pfam05, COG12, and cd0008)
- all three architectures (all three SPARCLE records) will be indexed with the same EC number that was annotated on SwissProt P0321
|
 3.6.4.13[ECNumber]
3.6.4.13[ECNumber]
will retrieve architectures that have the Enzyme Commission number of 3.6.4.13, RNA helicase.
|
| Filter |
[Filter] |
The [Filter] field can be used to limit your search to conserved domain architectures that have links to another Entrez database of interest, as shown in the search examples to the right.
NCBI uses the following methods to create links between conserved domain architectures and records in other databases:
The SPARCLE data processing pipeline calculates two types of direct links:
- sparcle_protein: each conserved domain architecture in the SPARCLE database links to all protein sequences that have the architecture.
- sparcle_cdd: each conserved domain architecture in the SPARCLE database links to all of the conserved domain models (specific hits and superfamilies) that compose the architecture. For example, if an architecture contains one specific hit and one superfamily, that SPARCLE record will link to two Conserved Domain Database (CDD) records -- one for the specific hit and one for the superfamily.
All other links between SPARCLE and other Entrez databases are indirect, created by a join between the proteins that contain the architecture and the other data types.
- For example, links from SPARCLE architectures to Gene records are created by a join between the following:
sparcle_protein AND protein_gene → sparcle_gene
|
 "chloride channel"[All] AND "sparcle_gene"[Filter]
"chloride channel"[All] AND "sparcle_gene"[Filter]
will retrieve conserved domain architectures that have the phrase "chloride channel" in any field of the record, and have links to records in the Gene database.
"chloride channel"[All] AND "sparcle_biosystems"[Filter]
will retrieve conserved domain architectures that have the phrase "chloride channel" in any field of the record, and have links to records in the Biosystems database.
(Note: To view the biosystems that are linked to an architecture, click on an architecture of interest in the SPARCLE search results, then click on the "pathways" link in the right hand margin of the architecture's summary page to open the corresponding Biosystems records.)
|
| GeneDescription |
[GeneDescription] |
The description of Gene records that were used as supporting evidence for conserved domain architectures.
The [GeneDescription] index includes text terms from the gene's official full name, official symbol, alternative symbols, and gene summary.
|
 "chloride channel"[GeneDescription]
"chloride channel"[GeneDescription]
will retrieve the architecture that lists, as supporting evidence, genes that include the phrase "chloride channel" in their description.
|
| GeneSymbol |
[GeneSymbol] |
The gene symbol of Gene records that were used as supporting evidence for conserved domain architectures. |
 nat16[GeneSymbol]
nat16[GeneSymbol]
will retrieve the architecture that lists, as supporting evidence, genes whose symbol is "nat16."
|
| Label |
[Label] |
The functional label (description) of a conserved domain architecture. |
 "chloride channel"[Label]
"chloride channel"[Label]
will retrieve the architecture(s) that contain the phrase "chloride channel" in the functional Label (description) of the architecture.
|
| Name |
[Name]
[NM] |
The name of a conserved domain architecture.
The data processing section of this document describes the three different methods by which conserved domain architectures are named:
- Curated architectures
- Autonamed architectures
- NamedByDomain architectures
These represent three tiers of SPARCLE records, which can be retrieved, if desired, using the [ReviewLevel] search field.
|
 "chloride channel"[Name]
"chloride channel"[Name]
will retrieve the architecture(s) that contain the phrase "chloride channel" in the name of the architecture.
|
| Organism |
[Organism]
[Orgn] |
The taxonomic node to which the name and label of the conserved domain architecture apply.
By default, conserved domain architectures are associated with the root of the taxonomic tree (i.e., all organisms). When an architecture is associated with the root, it means the name/label of the architecture is not specific to any node of the full taxonomic tree. This is true of most architectures in the SPARCLE database.
If the [Organism] classification of an architecture is not root, but is instead a more specific taxonomic node, that means the curator is asserting that the name/label chosen for the architecture is applicable within the specified node, but not necessarily within other taxonomic branches.
--------------------
For example, the total number of architectures in the SPARCLE database was 129405 as of July 13, 2017. (Note: the current total number of architectures might be larger or smaller, if more architectures have been added or removed since that date as a result of ongoing research).
Most of those architectures are assigned, by default, to the root of the taxonomic tree. (As an example, retrieve the architectures that have a taxonomic scope of all organisms.)
A small number of architectures are assigned to more specific taxonomic nodes, as follows:
The next column provides examples of search strategies that will retrieve conserved domain architectures that have a taxonomic scope of interest.
The SPARCLE record for each architecture contains a section entitled "Curated names and labels, which includes the architecture's taxonomic scope.
|
 bacteria[Organism]
bacteria[Organism]
will retrieve the architectures whose names and labels are applicable within bacteria but not within other taxonomic nodes.
viruses[Organism]
will retrieve the architectures whose names and labels are applicable within viruses but not within other taxonomic nodes.
guanylate cyclase AND bacteria[Organism]
will retrieve the architectures that contain the terms "guanylate" and "cyclase" in any field of the SPARCLE architecture record, and whose names and labels are applicable within bacteria but not within other taxonomic nodes.
guanylate cyclase AND eukaryota[Organism]
will retrieve the architectures that contain the terms "guanylate" and "cyclase" in any field of the SPARCLE architecture record, and whose names and labels are applicable within eukaryota but not within other taxonomic nodes.
|
| PDBTitle |
[PDBTitle]
[PDBTL] |
The title of any Protein Data Bank (PDB) record (3D macromolecular structure) that was used as supporting evidence for the conserved domain architecture. |
 "DNA modification"[PDBTitle]
"DNA modification"[PDBTitle]
will retrieve the SPARCLE record that contains the phrase "DNA modification" in the title of any 3D structure record that was used as supporting evidence for the conserved domain architecture. |
| ReviewLevel |
[ReviewLevel]
[REV] |
The SPARCLE database has three tiers (review levels) of conserved domain architecture records:
- Curated architectures
- Autonamed architectures
- NamedByDomain architectures
The data processing section of this document describes the methods by which architectures in each tier are handled.
The [ReviewLevel] search field can be used to limit retrieval to a specific tier of records, if desired, as shown in the search examples in the next column.
(Note: The [ReviewLevel] field is similar to the [Status] field, described below.)
|
 curated[ReviewLevel]
curated[ReviewLevel]
will retrieve all of the curated architectures from the SPARCLE database.
autonamed[ReviewLevel]
will retrieve all of the autonamed architectures from the SPARCLE database.
namedbydomain[ReviewLevel]
will retrieve all of the architectures from the SPARCLE database that were named by domain.
"chloride channel" AND curated[ReviewLevel]
will retrieve all architectures that contain the phrase "chloride channel" in any field of the record, and will then limit the retrieval to curated architectures.
|
| Status |
[Status] |
The [Status] field is similar to the [ReviewLevel] field (described above).
The [Status] field divides the SPARCLE database into two broad subsets:
- Reviewed (which represents curated records)
- Provisional (which represents all other SPARCLE records, such as those that were autonamed or namedByDomain)
(In contrast, the [ReviewLevel] field divides the SPARCLE database based on the method by which the data have been processed (i.e., curated, autonamed, namedByDomain).
Because of this, a search for curated[ReviewLevel] will retrieve the same subset of architectures as reviewed[Status].
A search for provisional[Status] will retrieve all architectures that have not been curated.
|
 reviewed[Status]
reviewed[Status]
will retrieve all of the reviewed (i.e., curated) architectures from the SPARCLE database.
"chloride channel" AND reviewed[Status]
will retrieve all architectures that contain the phrase "chloride channel" in any field of the record, and will then limit the retrieval to reviewed (i.e., curated) architectures.
|
| UID |
[UID]
[ArchID] |
The unique identification number (UID) of a conserved domain architecture. It is also referred to as an architecture ID, or archid.
If you enter an integer as a query, the search system will interpret the query by default as a search of the [UID] field.
Additional information about architecture IDs is provided in the section of this document that describes the contents of a conserved domain architecture's summary page.
|
 10087058[UID]
10087058[UID]
The search above, which uses the [UID] field specifier, will retrieve the architecture that has the unique identification number (UID) 10087058.
10087058
If you enter the query as just the integer, as shown above, without the [UID] field specifier, the search system will search the [UID] field by default.
Therefore, both of the searches above will retrieve the same architecture.
|
|
* In a query, the field name may be typed as the full name or abbreviation, and may be in upper, lower, or mixed case. If more than one abbreviation is shown, any one of them can be used. The field name must be surrounded by square brackets []. A space between the search term and the field specifier is optional. If desired, surround a phrase with quotes to force an adjacency search. For example, all of the sample queries below will work equally:
"chloride channel"[NAME]
"chloride channel" [NAME]
"chloride channel"[name]
"chloride channel" [name]
"chloride channel" [NM]
"chloride channel"[nm]
** The quotes surrounding the query terms in some of the sample searches force the terms to be searched as a phrase. If quotes are not used, the Entrez system may still recognize and handle the terms as a phrase, if they are present in a phrase dictionary used by the search engine. If the terms are not present in the phrase dictionary and are not surrounded by quotes, Entrez will insert a Boolean AND between the terms; in that case, they may or may not appear adjacent to each other in the retrieved records. The "Details" section in the right hand margin of a search results page will show you exactly how the Entrez system parsed your query. More search tips are provided in the PubMed help document and Entrez help document.
It is also possible to search for a word stem by using an asterisk (*) as a wild card; for example, arachidon* will retrieve records with terms such as arachidonate, arachidonic, arachidonoyl. The Entrez Help document provides additional information about truncating search terms in this way.
|



![Step 1 in searching the SPARCLE database by keyword: Enter the desired search terms in the query box, adding curated[ReviewLevel], if desired, to limit results to curated domain architectures. Click on this graphic to open the SPARCLE home and input your own search terms.](images/entrez_sparcle_step1_home_page_search_for_chloride_channel_curated_reviewlevel.png)
![Step 2 in searching the SPARCLE database by keyword: View the search results and click on the architecture ID of any domain architecture of interest to open its summary page. Click on this graphic to open the results of a SPARCLE search for chloride channel AND curated[ReviewLevel].](images/entrez_sparcle_step2_search_results_chloride_channel_curated_reviewlevel.png)
