All WormBook content, except where otherwise noted, is licensed under a Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
WormBook: The Online Review of C. elegans Biology [Internet]. Pasadena (CA): WormBook; 2005-2018.
There are two sexes in C. elegans, hermaphrodite and male. While there are many sex-specific differences between males and hermaphrodites that affect most tissues, the basic body plan and many of its structures are identical. However, most structures required for mating or reproduction are sexually dimorphic and are generated by sex-specific cell lineages. Thus to understand cell fate specification in hermaphrodites, one must consider how the body plan, which is specified during embryogenesis, influences the fates individual cells. One possible mechanism may involve the asymmetric distribution of POP-1/Tcf, the sole C. elegans Tcf homolog, to anterior-posterior sister cells. Another mechanism that functions to specify cell fates along the anterior-posterior body axis in both hermaphrodites and males are the Hox genes. Since most of the cell fate specifications that occur in hermaphrodites also occur in males, the focus of this chapter will be on those that only occur in hermaphrodites. This will include the cell fate decisions that affect the HSN neurons, ventral hypodermal P cells, lateral hypodermal cells V5, V6, and T; as well as the mesodermal M, Z1, and Z4 cells and the intestinal cells. Both cell lineage-based and cell-signaling mechanisms of cell fate specification will be discussed. Only two direct targets of the sex determination pathway that influence cell fate specification to produce hermaphrodite-specific cell fates have been identified. Thus a major challenge will be to learn additional mechanisms by which the sex determination pathway interacts with signaling pathways and other cell fate specification genes to generate hermaphrodite-specific cell fates.
1. Introduction
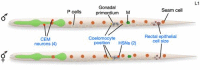
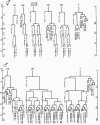
There are two sexes in C. elegans, hermaphrodite and male. Hermaphrodites are basically females that produce a small number of sperm that can fertilize their own oocytes. They are also cross-fertile with males. Males are smaller than hermaphrodites and produce only sperm. There are many sex-specific differences between males and hermaphrodites. Although these differences affect most tissues, the basic body plan and many of its structures are identical (Sulston et al., 1980; Sulston and Horvitz, 1977). In fact, about 650 cells seem to be the same in both sexes, out of a total of 959 somatic nuclei in hermaphrodites, and 1031 somatic nuclei in males. Most structures required for mating or reproduction are sexually dimorphic. For example, the hermaphrodite vulva is a reproductive structure that functions to allow eggs to be laid and male sperm to be deposited. Males develop highly specialized reproductive structures in the tail that allow it to find the hermaphrodite vulva and deposit sperm (Figure 1). Furthermore, the gonads of each sex have different structures and functions. Finally, sexually dimorphic neurons and muscles develop in each sex to control the movements of these mating structures.
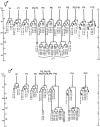
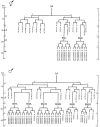
Each of these structures is generated by sex-specific cell lineages. For example, certain P cells divide to generate the vulva in hermaphrodites, while other P cells divide to generate portions of the mating apparatus of the male tail (Figure 2). Other male specific blast cells that do not divide in hermaphrodites generate additional mating structures in the tail. In addition, the M cell divides in hermaphrodites to generate the vulval muscles that control the movements of the vulva. In males, the M cell divides to generate sex muscles that control movements of the tail (Figure 3). Interestingly, almost all of these sexually dimorphic structures are made during larval development by the sex-specific divisions of just 16 blast cells (Figure 4).
Since most of the cell fate specifications that occur in hermaphrodites also occur in males, the focus of this chapter will be on those that only occur in hermaphrodites. Thus, I will include the cell fate decisions that affect the HSN neurons, ventral hypodermal P cells, lateral hypodermal cells V5, V6, and T; as well as the mesodermal M, Z1, and Z4 cells. Even the intestinal cells have sex-specific fates (Figure 5). Aspects of many of these cell fate decisions are covered in more detail in other chapters, which will be referred to where appropriate.
2. Basic body plan and role of Hox genes
The basic body plan of C. elegans is specified during embryogenesis through both cell interactions and autonomous or cell lineage-based mechanisms (see Asymmetric cell division and axis formation in the embryo, Gastrulation in C. elegans and Epidermal morphogenesis). The body plan must influence the fates of individual cells, however the mechanisms by which this occurs are not yet clear. One possible mechanism may involve POP-1/Tcf. POP-1 is the sole Tcf homolog in C. elegans. Tcfs are transcription factors that function to activate or repress expression of specific genes in response to Wnt signals (see Wnt chapter). POP-1 is asymmetrically distributed to anterior-posterior sister cells in many divisions during C. elegans development (Herman, 2002; Herman and Wu, 2004; Lin et al., 1998). At most divisions, the nucleus of the anterior daughter has a higher level of POP-1 than does the posterior daughter. Early in embryogenesis POP-1 asymmetry is generated by cell interactions that involve MOM-2/Wnt signals. However later, after the division of the AB great-granddaughter cells (AB8), the generation of POP-1 asymmetry becomes independent of MOM-2, yet still requires MOM-5/Frizzled (Park and Priess, 2003). Recently, it was shown that prior to POP-1 being asymmetric at many of these divisions, MOM-5 is asymmetrically distributed during the division, making this system reminiscent of the Drosophila and vertebrate planar cell polarity pathways (Park et al., 2004). Thus, the asymmetric distribution of POP-1/Tcf might be controlled by a planar cell polarity-like pathway and function as part of a "global patterning system" that may control many cell fates. However, there must be other mechanisms at work as well, since not all cell fates are defective in pop-1 mutants (Herman, 2001; Lin et al., 1998; Siegfried and Kimble, 2002). Furthermore, this system must function in both hermaphrodites and males.
Another mechanism that functions to specify cell fates along the anterior-posterior body axis in both hermaphrodites and males is the Hox genes. There are six Hox genes in the C. elegans genome. All are found within 5 Mb on chromosome III: four, ceh-13, lin-39, mab-5 and egl-5 are found in a loose cluster, whereas two others, nob-1 and php-3 are located more than 1 Mb away (Aboobaker and Blaxter, 2003; Burglin et al., 1989; Burglin and Ruvkun, 1993; Burglin et al., 1991; Ruvkun and Hobert, 1998; Van Auken et al., 2000). Interestingly, lin-39, mab-5 and egl-5 are only required for postembryonic development (Kenyon et al., 1997). Furthermore, it appears that unlike Drosophila or vertebrate systems, much of the Hox gene expression pattern in C. elegans is dictated by cell lineage (Cowing and Kenyon, 1996). However, cell signals such as the Wnt signaling pathways control some aspects of Hox gene expression (Streit et al., 2002), particularly in the case of vulval development (see Vulva development; Eisenmann et al., 1998). As in other systems, the Hox genes function to specify regions along the anterior-posterior body axis and, with the exception of ceh-13, their positions along the chromosome correlate with their regions of expression along the body axis. While mab-5 and egl-5 play large roles in cell fate specification in males (see Male development), in hermaphrodites, their roles are minimal (see below). lin-39 is a major determinant of vulval cell fates (see Vulval development). Interestingly, it has been proposed that a tissue polarity or PCP-like mechanism might specify the Hox expression pattern in C. elegans (Kenyon et al., 1997). It is possible that the two large-scale patterning mechanisms discussed here may interact to produce different cell fates along the body axis.
3. HSN neuron
Although programmed cell death plays a large role in shaping the body plan during development, it does not play a large role in shaping the differences between males and hermaphrodites. However, the HSN and CEM neurons are notable exceptions. The two serotonergic hermaphrodite specific neurons (HSNs) innervate the vulval muscles and drive egg-laying in hermaphrodites. The HSNs are not needed in males and die during embryonic development. By contrast, the four CEMs are male-specific head neurons that may be involved in regulating male-specific behaviors. Thus they are not needed in hermaphrodites and die during embryogenesis (Sulston and Horvitz, 1977; see Male development). HSN death in males is controlled by sex-specific expression of the pro-apoptotic gene, egl-1 by the terminal sex-determination gene tra-1 (Conradt and Horvitz, 1998). Gain-of-function mutations in egl-1 that cause inappropriate death of the HSNs in hermaphrodites (Desai et al., 1988; Trent et al., 1983) map to a TRA-1A binding site, indicating that TRA-1A functions as a sex-specific negative regulator of egl-1 expression (Conradt and Horvitz, 1999). Thus, the sex determination pathway directly controls sex-specific HSN fates. This is one of two known examples of control of somatic sexual fates by TRA-1A. The other example involves the control of yolk production in the intestine by mab-3 (see below).
4. Epidermal cell fates
In C. elegans (and other nematodes), the epidermis is referred to as hypodermis. Although the overall organization of the hypodermis in males and hermaphrodites is similar, there are many sex-specific specializations, including the development of the hermaphrodite vulva from the ventral hypodermis and the male sensory rays from the lateral hypodermis. At hatching, there are 85 hypodermal nuclei that are derived from only four blastomeres of the 12-cell embryo. The GATA transcription factor ELT-1 appears to be required for all hypodermal cell fates (Gilleard and McGhee, 2001; Page et al., 1997). Ectopic expression of ELT-1, or the related GATA factor, ELT-3, in early blastomeres, can activate the expression of hypodermal cell fate markers and cause cells that normally do not produce hypodermis to become hypodermal (Gilleard and McGhee, 2001). One of these markers is lin-26, which encodes a zinc finger transcription factor (Labouesse et al., 1994) that is expressed in all epithelial cells of the ectoderm as well as in the somatic gonad. In lin-26 mutants epidermal cells are produced, however they soon degenerate, suggesting that lin-26 is required for epithelial cell differentiation (den Boer et al., 1998; Labouesse et al., 1996). Furthermore, ectopic expression of LIN-26 in early embryonic blastomeres activates the expression of several other epithelial markers, suggesting that lin-26 is a major control gene for epithelial cell fates (Quintin et al., 2001). Recent studies of the lin-26 promoter revealed the existence of multiple regulatory elements that controlled expression in cells of the same organ, cells of similar function, or cells that had similar positions along the body axis (Landmann et al., 2004). Thus, LIN-26 functions to control epithelial cell differentiation in multiple lineages using modular promoter elements.
4.1. Lateral hypodermis
Three major cell types make up the C. elegans epidermis: the hypodermal syncytium that covers much of the body, the seam cells and the P cells, whose nuclei migrate to the ventral nerve cord (Figure 6). The GATA factors ELT-5 and ELT-6 are required for the seam cell fate. In the absence of elt-5 and elt-6 function, the seam cells inappropriately fuse with the hypodermal syncytium (Koh and Rothman, 2001). The seam cells are generated by divisions of the H, V and T cells (Figure 7). At hatching, there are eight seam cells along the each side of the animal, H1 and H2 in the head; V1-V6, along the body; and T in the tail. In wild-type hermaphrodites, the anterior V1-V4 cells and the posterior V6 cell all divide asymmetrically in a similar stem cell-like manner throughout larval development; the anterior daughter fuses with the hypodermal syncytium, hyp7, and the posterior daughter becomes a new seam cell. After the last division, all the posterior daughters differentiate into cells that produce the alae, which are cuticular ridges that run the length of the body (Sulston and Horvitz, 1977). The H cells also divide in a stem-like manner, but their division patterns differ from V1-V4 and from each other. The V5 cell also undergoes stem cell-like divisions, but differs from V1-V4 and V6 because the V5.pa cell becomes a neuroblast that generates a sensory structure called the postdeirid. When seam cells anterior or posterior to V5.p are killed with a laser microbeam, V5.pa fails to generate a postdeirid, suggesting that physical contact with V4.p and V6.p is required (Sulston and White, 1980). This signaling appears to occur after the first round of seam cell divisions, when contacts between V5.p and its neighbors are reestablished (Austin and Kenyon, 1994). Wnt signals appear to be responsible for the induction of postdeirid cell fates (see Wnt signaling). In males, the V5, V6 and T cells produce sensory rays in a process that involves Wnt signals and the regulation of Hox genes mab-5 and egl-5, as well as several cofactors (see Wnt signaling and Male development). The asymmetric divisions of two of these cells are also regulated by cell signals. Specifically, mutations in cam-1, which encodes a member of the Ror kinase family causes polarity reversal of the V1 cell division (Forrester et al., 1999); whereas mutations in egl-20/Wnt cause polarity reversal of the V5 cell division (Whangbo et al., 2000).
lin-22, which encodes a homolog of hairy/Enhancer of split, is also involved in specifying V cell fates (Wrischnik and Kenyon, 1997). In lin-22 mutants, V1-V4 adopt the fate of V5 (Fixsen et al., 1985; Horvitz et al., 1983; Wrischnik and Kenyon, 1997). Production of ectopic postdeirids in lin-22 mutants required the activities of mab-5, the achaete-scute homolog, lin-32, and to some extent lin-39; suggesting that LIN-22 functions to inhibit the functions of these proteins in V5.pa (Wrischnik and Kenyon, 1997). It is not yet clear how LIN-22 function is affected by the Wnt signals that appear to be repressed by cell contacts.
The T cells divide with a different pattern than the V cells and primarily generate neurons and neural support cells, although syncytial and seam cells are also produced. The asymmetric T cell divisions are controlled by a Wnt pathway that involves lin-44/Wnt, lin-17/Fz, wrm-1/β-catenin, lit-1/NLK and pop-1/Tcf (see Wnt signaling for details; Herman, 2001; Herman and Horvitz, 1994; Herman et al., 1995; Sawa et al., 1996). A likely target of this pathway is tlp-1, which encodes a zinc finger protein expressed in the posterior T cell daughter in a Wnt dependant manner and appears to function to specify neural cell fates (Zhao et al., 2002).
4.2. Ventral hypodermis
At hatching, the nuclei of six ventrolateral P cells lie along each side of the animal. About midway through the first larval stage the nuclei migrate into the ventral nerve cord, followed subsequently by the remaining cell body (Sulston et al., 1983). Laser killing experiments demonstrated that cells that lie opposite each other have equivalent cell fates. If one member of the pair is killed, the remaining member takes on the anterior cell fate (Sulston and White, 1980). Once in the ventral cord, the cells divide producing an anterior Pn.a neuroblast that generates several motorneurons and a posterior hypodermal Pn.p cell. In hermaphrodites, P(1–2).p and P(9–11).p soon fuse with hyp-7, while P(3–8).p remain unfused. In hermaphrodites P(3–8).p make-up the vulval precursor cells and participate in vulval development during the third larval stage (see Vulva development chapter for details). In males, P(1–2).p and P(7–8).p soon fuse with hyp7, while P(3–6).p and P(9–11).p remain unfused. P12.p also remains unfused and divides in the first larval stage with P12.pa generating a rectal hypodermal cell and P12.pp undergoing programmed cell death in both sexes. The pattern of Pn.p fusions, and thus vulval competency in hermaphrodites, appears to be controlled by Hox gene expression and Hox protein activity. In both wild-type hermaphrodites and males, lin-39 is expressed in P(3–8).p and mab-5 is expressed in P(7–11).p (Maloof and Kenyon, 1998; Salser et al., 1993). However, the activity of MAB-5 is inhibited in hermaphrodites, perhaps by the action of the sex determination pathway (Salser et al., 1993), although a direct role for TRA-1A has not been shown. egl-27, which encodes a protein similar to the NURD chromatin remodeling complex component MTA1, is also required for the MAB-5 inhibition in hermaphrodites (Herman et al., 1999; Solari et al., 1999). In males, MAB-5 and LIN-39 neutralize each other in a combinatorial interaction that also requires egl-27 function, leading to the fusion of P(7–8).p with hyp7 (Ch'ng and Kenyon, 1999).
The fate of the P12 cell is controlled by an interaction of Wnt and Ras pathways that interact to regulate the expression of the Hox gene egl-5 (see Wnt signaling).
4.3. M cell
Although most of the main body wall muscles are not sexually dimorphic (Hodgkin, 1988; Sulston et al., 1980; Sulston et al., 1980; Sulston and Horvitz, 1977), the single postembryonic mesoblast, M, produces different muscles in hermaphrodites and males. M, a descendant of MS.ap, is born in an anterior position on the left side of the embryo. It then migrates to the posterior and makes its way to the right side of the embryo, where it is found after hatching (Figure 1; Sulston et al., 1983). In hermaphrodites, the M cell produces six cell types: body wall muscles (14), nonmuscle coelomocytes (2) and four classes of sex muscles (4 cells of each class), which are derived from two sex myoblast (SM) cells (Figure 3). The SMs are born in the posterior and migrate toward the gonad during the second larval stage where they divide in the third larval stage to generate the uterine (um1 and um2) and vulval (vm1 and vm2) muscles. This migration is controlled by both attractive and repulsive signals from the gonad (Stern and Horvitz, 1991; Thomas et al., 1990). The attractive signals involve the expression of EGL-17/FGF by the somatic gonad and vulva and EGL-15/FGFr by the SMs. In the absence of egl-15 or egl-17 function, a repressive signal from the gonad is revealed. The function and exact source of this signal within the gonad is not yet known (Branda and Stern, 2000; Burdine et al., 1998; Stern and Horvitz, 1991).
Multiple regulatory factors have been shown to be involved in the proliferation and patterning of the M lineage, as well as for cell fate decisions between body wall muscles, coelomocytes and sex myoblasts. These include lin-12/Notch (Greenwald et al., 1983), mab-5 (Kenyon, 1986), hlh-1/MyoD (Harfe et al., 1998; Krause et al., 1990), hlh-8/twist (Harfe et al., 1998; Liu and Fire, 2000) and unc-39/Six4/5 (Yanowitz et al., 2004). Hox genes lin-39 and mab-5 play overlapping roles in specifying the various M cell derived fates through the activation of several targets (Liu and Fire, 2000). One of the main Hox gene targets is hlh-8/twist which is expressed in undifferentiated M cell descendants. Mutations in hlh-8 cause both the striated and nonstriated M-derived muscles to be misspecified (Harfe et al., 1998). HLH-8 functions with HLH-2 to activate additional target genes. hlh-2 encodes an E/Daughterless homolog and is expressed in a dynamic pattern throughout development, including in the M lineage (Krause et al., 1997). After the initial M cell divisions, hlh-1 and lin-12 function to specify striated muscle, coelomocyte and sex myoblast fates. Mutations in hlh-1 cause the coelomocytes to be transformed into sex myoblasts, while mutations in lin-12 cause the opposite transformation (Greenwald et al., 1983; Harfe et al., 1998; Krause et al., 1990).
A genetic screen for mutations that affected cell fate specification within the SM lineage identified mls-1, which encodes a T-box transcription factor. Uterine muscles are transformed into vulval muscles in mls-1 mutants. Interestingly, overexpression of mls-1 in the M lineage causes the opposite transformation, indicating that MLS-1 is a uterine muscle cell fate determinant (Kostas and Fire, 2002).
5. Somatic gonad
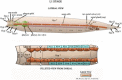
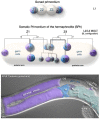
The gonads of hermaphrodites and males develop sex-specific morphologies from a common primordium. The gonad primordium lies in the center of the animal and contains four cells: two somatic gonadal precursors (SGPs), Z1 and Z4, on each end, and two germline precursors, Z2 and Z3, in the center. The hermaphrodite gonad develops an anterior and a posterior arm, each of which has its own proximal-distal axis and develops into an ovo-testes (Figure 8; see Introduction to the germ line). The male gonad develops into a single-armed testis (see Male development). Thus, the SGPs undergo sex-specific cell lineages (Kimble and Hirsh, 1979). The hermaphrodite SGPs divide along the proximal-distal axis of the gonad with distal cell fates lying at the ends of the primordium and proximal fates lying in the center. The SGPs divide three times in the first larval stage to generate the 12-cell gonad primordium (Figure 8). Ten of these cells have invariant fates, eight later generate uterine, sheath and spermathecal cells and two become the distal tip cells (DTCs) on each end which lead the elongation of the gonad arms. The remaining two cells, Z1.ppp and Z4.aaa have variable cell fates. One will become the anchor cell (AC) and the other will become a ventral uterine precursor (VU). However, which cell becomes the AC and which becomes the VU varies from animal to animal (Kimble and Hirsh, 1979).
A Wnt signaling pathway controls the asymmetric cell divisions of the SGPs. These asymmetric cell divisions and the resulting tissues are oriented along the proximal-distal axis of the gonad. Mutations in lin-17/Fz also cause a loss of asymmetry of the SGP divisions (Sternberg and Horvitz, 1988). In the gonad, this defect is called Sys (for symmetric sisters). Mutations in pop-1/Tcf also cause a Sys defect and POP-1 levels appear to be correlated with different proximal and distal cell fates, as the nuclear levels of GFP::POP-1 levels are higher in the proximal Z1/Z4 daughters (Siegfried et al., 2004; Siegfried and Kimble, 2002). The Wnt pathway that controls SGP polarity also includes mom-1/Porc, lit-1/NLK and wrm-1/β-catenin. Other recently identified Sys genes include sys-1, sys-3, gon-14, gon-1 and gon-16 (Siegfried et al., 2004). TRA-1 also controls SGP position and polarity, including the nuclear levels of GFP::POP-1 (Mathies et al., 2004). However, it is not clear how the sex determination pathway might interact with the Wnt signaling pathway to control SGP polarity. Surprisingly, mutation or interference with each of the C. elegans Wnts did not cause a Sys defect, thus the Wnt ligand involved in this asymmetric division remains unidentified. It is possible that two or more of the five C. elegans Wnts function redundantly to control SGP polarities, or that a non-ligand mediated mechanism, such as planar cell polarity, may be at work. However, if this is the case, there must be a mechanism by which the gonad can develop a proximal-distal axis of polarity that is different from the anterior-posterior axis of the animal.
The AC/VU cell fate decision is controlled by cell interactions mediated by the LIN-12/Notch receptor and LAG-2, which is a ligand of the Delta-Serrate-LAG-2 family. Initially, Z1.ppp and Z4.aaa express both lin-12 and lag-2, but a feedback mechanism functions to amplify a small fluctuation in lin-12 levels, causing one cell to express higher levels of lin-12 and stop transcribing lag-2, while the other cell begins to express higher levels of lag-2 and stops transcribing lin-12. As a result, the lin-12 expressing cell becomes a VU and the lag-2 expressing cell becomes the AC (for details see LIN-12/Notch signaling in C. elegans, Greenwald et al., 1983; Lambie and Kimble, 1991; Seydoux and Greenwald, 1989; Wilkinson et al., 1994). Recently it has been shown that HLH-2 is required for lag-2 expression during the AC/VU decision (Karp and Greenwald, 2003) as well as for establishing the competence of Z1.ppp and Z4.aaa to participate in the AC/VU decision (Karp and Greenwald, 2004).
6. Intestine
The hermaphrodite intestine produces yolk proteins, while the male intestine does not. This sex-specific cell fate is controlled by mab-3 and tra-1. MAB-3 is a DM domain containing transcription factor related to Drosophila Doublesex and mouse Dmrt1 (Raymond et al., 1998). Mutations in mab-3 cause yolk to be produced in males (Shen and Hodgkin, 1988). MAB-3 binds to a site in the promoter of the vit-2 vitellogenin gene in males, repressing vit-2 transcription (Yi et al., 2000). TRA-1A binds to a site in the mab-3 promoter in hermaphrodites, repressing mab-3 transcription (Yi and Zarkower, 1999). Thus, the hermaphrodite specific fate of the intestinal cells is controlled directly by the sex determination pathway.
7. Conclusions
Although there do not appear to be any general genetic or developmental strategies that differentiate hermaphrodite from male development, some trends are evident. Sex-specific differences in the patterns of programmed cell death appear to play a minor role, whereas sex-specific differences in the patterns of cell division play a large role. Specifically, there are few examples of sex-specific programmed cell deaths: the HSNs die in males whereas the CEMs and the progenitors of the CA/P(1–6) neurons die in hermaphrodites. Sex-specific divisions of larval blast cells are more common. One result of this is that newly hatched larvae of each sex have virtually identical morphologies whereas adult hermaphrodites and males have unique appearances. In some cases divide blast cells in one sex but not in the other. For example the B and F cells divide in males, but not in hermaphrodites. In other cases blast cells divide in both sexes, but have sex specific cell lineages. For example the Pn.p and Vn.p cells divide differently in hermaphrodites and males. Finally, in some cases, such as the intestine, sex-specific gene expression is important but does not lead to differences in morphology of the tissue. A detailed understanding of how cell fates are differentially specified will go a long way to understanding the control of sex-specific development.
Many C. elegans papers on cell lineage or cell fate specification that were published in the late 1970s and 1980s emphasized the invariance of the cell lineage (Deppe et al., 1978; Kimble and Hirsh, 1979; Sulston and Horvitz, 1977; Sulston et al., 1983 and 97 others), suggesting that cell fates were primarily determined by cell intrinsic or lineage-based mechanisms. However, it was know quite early on that there were many examples of cell fate determination by cell interactions (Kimble, 1981; Sulston and White, 1980). Subsequent work has demonstrated that while many cell fates do indeed appear to be specified by lineage-based mechanisms, there is a richness of cell signaling systems also at work. A major future challenge will be to identify and understand the global mechanisms that may function to specify cell positions and cell fates along the body axis. As suggested earlier in this chapter, this may involve a linage-based system involving POP-1/Tcf and the interaction of multiple signaling pathways. A continuing challenge is to understand how lineage-based mechanisms and signaling systems interact to specify cell fates.
An additional challenge will be to understand how sex specific cell fates are specified. How does the pathway described in the Somatic sex determination chapter interact with the various cell fate determination pathways described here and elsewhere in this book to generate sex specific patterns of cell division, morphology and function? The mechanisms by which the sex determination pathway interacts with signaling pathways such as the Wnt pathway in the control of SGP polarity and fate are not yet clear. To date, only two direct somatic targets of TRA-1A, egl-1 and mab-3, have been identified. In both cases TRA-1A represses transcription of these targets in males. How does TRA-1A control the sex-specific fates in other cells? Genome analysis has revealed four genes in addition to mab-3 that have three or more TRA-1A upstream binding sties (Clarke and Berg, 1998). Three of these genes were enriched in expression in males vs. hermaphrodites (Jiang et al., 2001). However, egl-1 only has one TRA-1A binding site, which is located downstream, so it is unclear how many other genes may be directly regulated by TRA-1. Finally, it is likely that other downstream effectors will also interact with cell fate specification genes to control sex specific fates. The mechanisms by which the sex determination pathway interacts with other signaling pathways to control sex-specific cell fates remain to be discovered.
8. Acknowledgements
Work on C. elegans development in the Herman lab is supported by NIH grant GM56339. This publication was also made possible by NIH grant P20 RR016475 from the INBRE Program of the National Center for Research Resources.
9. References
- Aboobaker A. , Blaxter M. Hox gene evolution in nematodes: novelty conserved. Curr. Opin. Genet Dev. (2003);13:593–598. [PubMed: 14638320]
- Austin J. , Kenyon C. Cell contact regulates neuroblast formation in the Caenorhabditis elegans lateral epidermis. Development. (1994);120:313–323. [PubMed: 8149911]
- Branda C.S. , Stern M.J. Mechanisms controlling sex myoblast migration in Caenorhabditis elegans hermaphrodites. Dev. Biol. (2000);226:137–151. [PubMed: 10993679]
- Burdine R.D. , Branda C.S. , Stern M.J. EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans. Development. (1998);125:1083–1093. [PubMed: 9463355]
- Burglin T.R. , Finney M. , Coulson A. , Ruvkun G. Caenorhabditis elegans has scores of homoeobox-containing genes. Nature. (1989);341:239–243. [PubMed: 2571091]
- Burglin T.R. , Ruvkun G. The Caenorhabditis elegans homeobox gene cluster. Curr. Opin. Genet Dev. (1993);3:615–620. [PubMed: 7902148]
- Burglin T.R. , Ruvkun G. , Coulson A. , Hawkins N.C. , McGhee J.D. , Schaller D. , Wittmann C. , Muller F. , Waterston R.H. Nematode homeobox cluster. Nature. (1991);351:703. [PubMed: 1676487]
- Ch'ng Q. , Kenyon C. egl-27 generates anteroposterior patterns of cell fusion in C. elegans by regulating Hox gene expression and Hox protein function. Development. (1999);126:3303–3312. [PubMed: 10393110]
- Clarke N.D. , Berg J.M. Zinc fingers in Caenorhabditis elegans: finding families and probing pathways. Science. (1998);282:2018–2022. [PubMed: 9851917]
- Conradt B. , Horvitz H.R. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. (1998);93:519–529. [PubMed: 9604928]
- Conradt B. , Horvitz H.R. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell. (1999);98:317–327. [PubMed: 10458607]
- Cowing D. , Kenyon C. Correct Hox gene expression established independently of position in Caenorhabditis elegans. Nature. (1996);382:353–356. [PubMed: 8684464]
- den Boer B.G. , Sookhareea S. , Dufourcq P. , Labouesse M. A tissue-specific knock-out strategy reveals that lin-26 is required for the formation of the somatic gonad epithelium in Caenorhabditis elegans. Development. (1998);125:3213–3224. [PubMed: 9671593]
- Deppe U. , Schierenberg E. , Cole T.S. , Krieg C. , Schmitt D. , Yoder B. , Von E.G. Cell lineages of embryo of nematode C. elegans. Proc. Natl. Acad. Sci. USA. (1978);75:376–380. [PMC free article: PMC411251] [PubMed: 272653]
- Desai C. , Garriga G. , McIntire S.L. , Horvitz H.R. A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature. (1988);336:638–646. [PubMed: 3200316]
- Eisenmann D.M. , Maloof J.N. , Simske J.S. , Kenyon C. , Kim S.K. The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development. (1998);125:3667–3680. [PubMed: 9716532]
- Fixsen W. , Sternberg P. , Ellis H. , Horvitz H.R. Genes that affect cell fates during the development of Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. (1985);50:99–104. [PubMed: 3868513]
- Forrester W.C. , Dell M. , Perens E. , Garriga G. A C. elegans Ror receptor tyrosine kinase regulates cell motility and asymmetric cell division. Nature. (1999);400:881–885. [PubMed: 10476968]
- Gilleard J.S. , McGhee J.D. Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Mol. Cell. Biol. (2001);21:2533–2544. [PMC free article: PMC86885] [PubMed: 11259601]
- Greenwald I.S. , Sternberg P.W. , Horvitz H.R. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. (1983);34:435–444. [PubMed: 6616618]
- Harfe B.D. , Branda C.S. , Krause M. , Stern M.J. , Fire A. MyoD and the specification of muscle and non-muscle fates during postembryonic development of the C. elegans mesoderm. Development. (1998);125:2479–2488. [PubMed: 9609831]
- Harfe B.D. , Vaz Gomes A. , Kenyon C. , Liu J. , Krause M. , Fire A. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. (1998);12:2623–2635. [PMC free article: PMC317087] [PubMed: 9716413]
- Herman M. C. elegans POP-1/TCF functions in a canonical Wnt pathway that controls cell migration and in a noncanonical Wnt pathway that controls cell polarity. Development. (2001);128:581–590. [PubMed: 11171341]
- Herman M.A. Control of cell polarity by noncanonical Wnt signaling in C. elegans. Semin. Cell Dev. Biol. (2002);13:233–241. [PubMed: 12137732]
- Herman M.A. , Ch'ng Q. , Hettenbach S.M. , Ratliff T.M. , Kenyon C. , Herman R.K. EGL-27 is similar to a metastasis-associated factor and controls cell polarity and cell migration in C. elegans. Development. (1999);126:1055–1064. [PubMed: 9927605]
- Herman M.A. , Horvitz H.R. The Caenorhabditis elegans gene lin-44 controls the polarity of asymmetric cell divisions. Development. (1994);120:1035–1047. [PubMed: 8026318]
- Herman M.A. , Vassilieva L.L. , Horvitz H.R. , Shaw J.E. , Herman R.K. The C. elegans gene lin-44, which controls the polarity of certain asymmetric cell divisions, encodes a Wnt protein and acts cell nonautonomously. Cell. (1995);83:101–110. [PubMed: 7553861]
- Herman M.A. , Wu M. Noncanonical Wnt signaling pathways in C. elegans converge on POP-1/TCF and control cell polarity. Front Biosci. (2004);9:1530–1539. [PubMed: 14977564]
- Hodgkin, J. (1988) Sexual dimorphism and sex determination. In The Nematode Caenorhabditis elegans, W.B. Wood and the community of C. elegans researchers, eds., Cold Spring Harbor, NY, Cold Spring Harbor Laboratory, pp. 243–279.
- Horvitz H.R. , Sternberg P.W. , Greenwald I.S. , Fixsen W. , Ellis H.M. Mutations that affect neural cell lineages and cell fates during the development of the nematode Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. (1983);48(Pt 2):453–463. [PubMed: 6586368]
- Hubbard E.J. , Greenstein D. The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev. Dyn. (2000);218:2–22. [PubMed: 10822256]
- Jiang M. , Ryu J. , Kiraly M. , Duke K. , Reinke V. , Kim S.K. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. (2001);98:218–223. [PMC free article: PMC14571] [PubMed: 11134517]
- Karp X. , Greenwald I. Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans. Genes Dev. (2003);17:3100–3111. [PMC free article: PMC305261] [PubMed: 14701877]
- Karp X. , Greenwald I. Multiple roles for the E/Daughterless ortholog HLH-2 during C. elegans gonadogenesis. Dev. Biol. (2004);272:460–469. [PubMed: 15282161]
- Kenyon C. A gene involved in the development of the posterior body region of C. elegans. Cell. (1986);46:477–487. [PubMed: 3731276]
- Kenyon C.J. , Austin J. , Costa M. , Cowing D.W. , Harris J.M. , Honigberg L. , Hunter C.P. , Maloof J.N. , Muller-Immergluck M.M. , Salser S.J. et al. The dance of the Hox genes: patterning the anteroposterior body axis of Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. (1997);62:293–305. [PubMed: 9598363]
- Kimble J. , Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. (1979);70:396–417. [PubMed: 478167]
- Kimble J.E. Alterations in cell lineage following laser ablation of cells in the somatic gonad of C. elegans. Dev. Biol. (1981);87:286–300. [PubMed: 7286433]
- Koh K. , Rothman J.H. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development. (2001);128:2867–2880. [PubMed: 11532911]
- Kostas S.A. , Fire A. The T-box factor MLS-1 acts as a molecular switch during specification of nonstriated muscle in C. elegans. Genes Dev. (2002);16:257–269. [PMC free article: PMC155317] [PubMed: 11799068]
- Krause M. , Fire A. , Harrison S.W. , Priess J. , Weintraub H. CeMyoD accumulation defines the body wall muscle cell fate during C. elegans embryogenesis. Cell. (1990);63:907–919. [PubMed: 2175254]
- Krause M. , Park M. , Zhang J.M. , Yuan J. , Harfe B. , Xu S.Q. , Greenwald I. , Cole M. , Paterson B. , Fire A. A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development. (1997);124:2179–2189. [PubMed: 9187144]
- Labouesse M. , Hartwieg E. , Horvitz H.R. The Caenorhabditis elegans LIN-26 protein is required to specify and/or maintain all non-neuronal ectodermal cell fates. Development. (1996);122:2579–2588. [PubMed: 8787733]
- Labouesse M. , Sookhareea S. , Horvitz H.R. The Caenorhabditis elegans gene lin-26 is required to specify the fates of hypodermal cells and encodes a presumptive zinc-finger transcription factor. Development. (1994);120:2359–2368. [PubMed: 7956818]
- Lambie E.J. , Kimble J. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development. (1991);112:231–240. [PubMed: 1769331]
- Landmann F. , Quintin S. , Labouesse M. Multiple regulatory elements with spatially and temporally distinct activities control the expression of the epithelial differentiation gene lin-26 in C. elegans. Dev. Biol. (2004);265:478–490. [PubMed: 14732406]
- Lin R. , Hill R.J. , Priess J.R. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell. (1998);92:229–239. [PubMed: 9458047]
- Liu J. , Fire A. Overlapping roles of two Hox genes and the exd ortholog ceh-20 in diversification of the C. elegans postembryonic mesoderm. Development. (2000);127:5179–5190. [PubMed: 11060243]
- Maloof J.N. , Kenyon C. The Hox gene lin-39 is required during C. elegans vulval induction to select the outcome of Ras signaling. Development. (1998);125:181–190. [PubMed: 9486792]
- Mathies L.D. , Schvarzstein M. , Morphy K.M. , Blelloch R. , Spence A.M. , Kimble J. TRA-1/GLI controls development of somatic gonadal precursors in C. elegans. Development. (2004);131:4333–4343. [PubMed: 15294864]
- Page B.D. , Zhang W. , Steward K. , Blumenthal T. , Priess J.R. ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes Dev. (1997);11:1651–1661. [PubMed: 9224715]
- Park F.D. , Priess J.R. Establishment of POP-1 asymmetry in early C. elegans embryos. Development. (2003);130:3547–3556. [PubMed: 12810601]
- Park F.D. , Tenlen J.R. , Priess J.R. C. elegans MOM-5/frizzled functions in MOM-2/Wnt-independent cell polarity and is localized asymmetrically prior to cell division. Curr. Biol. (2004);14:2252–2258. [PubMed: 15620652]
- Quintin S. , Michaux G. , McMahon L. , Gansmuller A. , Labouesse M. The Caenorhabditis elegans gene lin-26 can trigger epithelial differentiation without conferring tissue specificity. Dev. Biol. (2001);235:410–421. [PubMed: 11437447]
- Raymond C.S. , Shamu C.E. , Shen M.M. , Seifert K.J. , Hirsch B. , Hodgkin J. , Zarkower D. Evidence for evolutionary conservation of sex-determining genes. Nature. (1998);391:691–695. [PubMed: 9490411]
- Ruvkun G. , Hobert O. The taxonomy of developmental control in Caenorhabditis elegans. Science. (1998);282:2033–2041. [PubMed: 9851920]
- Salser S.J. , Loer C.M. , Kenyon C. Multiple HOM-C gene interactions specify cell fates in the nematode central nervous system. Genes Dev. (1993);7:1714–1724. [PubMed: 8103754]
- Sawa H. , Lobel L. , Horvitz H.R. The Caenorhabditis elegans gene lin-17, which is required for certain asymmetric cell divisions, encodes a putative seven-transmembrane protein similar to the Drosophila frizzled protein. Genes Dev. (1996);10:2189–2197. [PubMed: 8804313]
- Seydoux G. , Greenwald I. Cell autonomy of lin-12 function in a cell fate decision in C. elegans. Cell. (1989);57:1237–1245. [PubMed: 2736627]
- Shen M.M. , Hodgkin J. mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell. (1988);54:1019–1031. [PubMed: 3046751]
- Siegfried K.R. , Kidd A.R. , 3rd Chesney , M.A. and , Kimble J. The sys-1 and sys-3 genes cooperate with Wnt signaling to establish the proximal-distal axis of the Caenorhabditis elegans gonad. Genetics. (2004);166:171–186. [PMC free article: PMC1470708] [PubMed: 15020416]
- Siegfried K.R. , Kimble J. POP-1 controls axis formation during early gonadogenesis in C. elegans. Development. (2002);129:443–453. [PubMed: 11807036]
- Solari F. , Bateman A. , Ahringer J. The Caenorhabditis elegans genes egl-27 and egr-1 are similar to MTA1, a member of a chromatin regulatory complex, and are redundantly required for embryonic patterning. Development. (1999);126:2483–2494. [PubMed: 10226007]
- Stern M.J. , Horvitz H.R. A normally attractive cell interaction is repulsive in two C. elegans mesodermal cell migration mutants. Development. (1991);113:797–803. [PubMed: 1821851]
- Sternberg P.W. , Horvitz H.R. lin-17 mutations of Caenorhabditis elegans disrupt certain asymmetric cell divisions. Dev. Biol. (1988);130:67–73. [PubMed: 3181641]
- Streit A. , Kohler R. , Marty T. , Belfiore M. , Takacs-Vellai K. , Vigano M.A. , Schnabel R. , Affolter M. , Muller F. Conserved regulation of the Caenorhabditis elegans labial/Hox1 gene ceh-13. Dev. Biol. (2002);242:96–108. [PubMed: 11820809]
- Sulston J.E. , Albertson D.G. , Thomson J.N. The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev. Biol. (1980);78:542–576. [PubMed: 7409314]
- Sulston J.E. , Horvitz H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. (1977);56:110–156. [PubMed: 838129]
- Sulston J.E. , Schierenberg E. , White J.G. , Thomson J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. (1983);100:64–119. [PubMed: 6684600]
- Sulston J.E. , White J.G. Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Dev. Biol. (1980);78:577–597. [PubMed: 7190941]
- Thomas J.H. , Stern M.J. , Horvitz H.R. Cell interactions coordinate the development of the C. elegans egg-laying system. Cell. (1990);62:1041–1052. [PubMed: 2401010]
- Trent C. , Tsuing N. , Horvitz H.R. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics. (1983);104:619–647. [PMC free article: PMC1202130] [PubMed: 11813735]
- Van Auken K. , Weaver D.C. , Edgar L.G. , Wood W.B. Caenorhabditis elegans embryonic axial patterning requires two recently discovered posterior-group Hox genes. Proc. Natl. Acad. Sci. USA. (2000);97:4499–4503. [PMC free article: PMC18263] [PubMed: 10781051]
- Whangbo J. , Harris J. , Kenyon C. Multiple levels of regulation specify the polarity of an asymmetric cell division in C. elegans. Development. (2000);127:4587–4598. [PubMed: 11023862]
- Wilkinson H.A. , Fitzgerald K. , Greenwald I. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell. (1994);79:1187–1198. [PubMed: 8001154]
- Wrischnik L.A. , Kenyon C.J. The role of lin-22, a hairy/enhancer of split homolog, in patterning the peripheral nervous system of C. elegans. Development. (1997);124:2875–2888. [PubMed: 9247331]
- Yanowitz J.L. , Shakir M.A. , Hedgecock E. , Hutter H. , Fire A.Z. , Lundquist E.A. UNC-39, the C. elegans homolog of the human myotonic dystrophy-associated homeodomain protein Six5, regulates cell motility and differentiation. Dev. Biol. (2004);272:389–402. [PubMed: 15282156]
- Yi W. , Ross J.M. , Zarkower D. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development. (2000);127:4469–4480. [PubMed: 11003845]
- Yi W. , Zarkower D. Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms. Development. (1999);126:873–881. [PubMed: 9927589]
- Zhao X. , Yang Y. , Fitch D.H. , Herman M.A. TLP-1 is an asymmetric cell fate determinant that responds to Wnt signals and controls male tail tip morphogenesis in C. elegans. Development. (2002);129:1497–1508. [PubMed: 11880358]
Footnotes
- *
Edited by: Barbara J. Meyer. Last revised August 18, 2005. Published January 09, 2006. This chapter should be cited as: Herman, M. A. Hermaphrodite cell-fate specification (January 09, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.39.1, http://www
.wormbook.org [PMC free article: PMC4781446] [PubMed: 18050480].
- §
To whom correspondence should be addressed. E-mail: ude.usk@namrehm
- The Paired-box protein PAX-3 regulates the choice between lateral and ventral epidermal cell fates in C. elegans.[Dev Biol. 2016]The Paired-box protein PAX-3 regulates the choice between lateral and ventral epidermal cell fates in C. elegans.Thompson KW, Joshi P, Dymond JS, Gorrepati L, Smith HE, Krause MW, Eisenmann DM. Dev Biol. 2016 Apr 15; 412(2):191-207. Epub 2016 Mar 4.
- The mab-21 gene of Caenorhabditis elegans encodes a novel protein required for choice of alternate cell fates.[Development. 1995]The mab-21 gene of Caenorhabditis elegans encodes a novel protein required for choice of alternate cell fates.Chow KL, Hall DH, Emmons SW. Development. 1995 Nov; 121(11):3615-26.
- Symmetry breakage in the development of one-armed gonads in nematodes.[Development. 1996]Symmetry breakage in the development of one-armed gonads in nematodes.Félix MA, Sternberg PW. Development. 1996 Jul; 122(7):2129-42.
- Review Cell fate specification and differentiation in the nervous system of Caenorhabditis elegans.[Dev Genet. 1996]Review Cell fate specification and differentiation in the nervous system of Caenorhabditis elegans.Sengupta P, Bargmann CI. Dev Genet. 1996; 18(1):73-80.
- Review Male development.[WormBook. 2005]Review Male development.Emmons SW. WormBook. 2005 Nov 10; :1-22. Epub 2005 Nov 10.
- Hermaphrodite cell-fate specification - WormBookHermaphrodite cell-fate specification - WormBook
Your browsing activity is empty.
Activity recording is turned off.
See more...